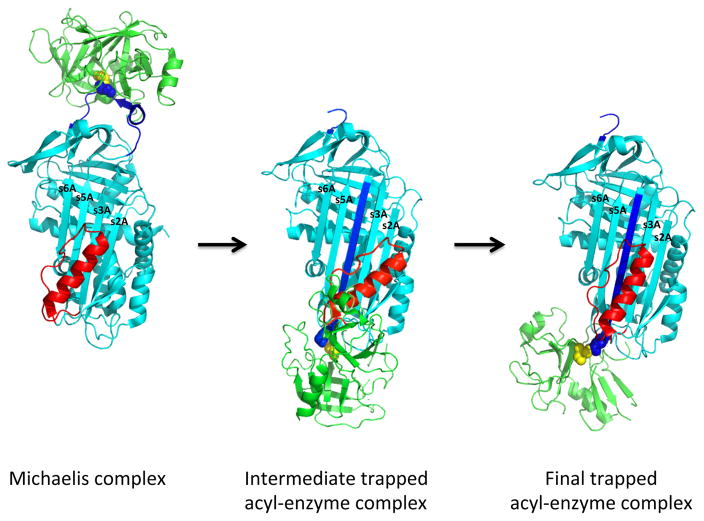
Fig 7. Two-stage mechanism for helix F participation in kinetic trapping of the acyl-enzyme complex.
The scheme depicts the hypothesized two-stage mechanism by which the serpin and proteinase conformational changes triggered by proteinase cleavage of the serpin RCL in the acyl-enzyme complex are coupled to kinetically trap the acyl-enzyme complex following its formation from the initial Michaelis docking complex. In the scheme, the Michaelis docking complex (serpin colored cyan with P1 residue in blue spheres and proteinase colored green with active-site Ala195 residue in yellow spheres) undergoes transformation to an intermediate conformationally altered acyl-enzyme complex in which the cleaved RCL (blue) has fully inserted into β-sheet A, with displacement of helix F (red) from its native position to allow translocation of the RCL-linked proteinase to an intermediate position at the s2A end of β-sheet A. In the intermediate, the acyl-linkage between the serpin P1 residue and the proteinase catalytic Ser195 (yellow spheres) is partially distorted and the F helix is constrained in a higher energy state. The intermediate is then transformed to the final trapped complex by the relaxation of the F helix to its unconstrained position with the energy released by this relaxation driving the concomitant movement of the proteinase to the s5A side of sheet A and causing full inactivation of the proteinase by conformational distortion. The initial and final complexes are x- ray structures of Michaelis (1OPH [180]) and trapped acyl-enzyme (1EZX [29]) complexes of α1PI with trypsin, whereas the intermediate complex is modeled based on FRET and fluorescence perturbation changes accompanying the reaction of fluorophore-labeled α1PIs with trypsin as described in the text.