Abstract
Randomized controlled trials can benefit from proactive assessment of how well their participant selection strategies during the design of eligibility criteria can influence the study generalizability. In this paper, we present a quantitative metric called generalizability index for study traits 2.0 (GIST 2.0) to assess the a priori generalizability (based on population representativeness) of a clinical trial by accounting for the dependencies among multiple eligibility criteria. The metric was evaluated on 16 sepsis trials identified from ClinicalTrials.gov, with their adverse event reports extracted from the trial results section. The correlation between GIST scores and adverse events was analyzed. We found that the GIST 2.0 score was significantly correlated with total adverse events and serious adverse events (weighted correlation coefficients of 0.825 and 0.709, respectively, with P < 0.01). This study exemplifies the promising use of Big Data in electronic health records and ClinicalTrials.gov for optimizing eligibility criteria design for clinical studies.
Keywords: clinical trials, generalizability, population representativeness, eligibility criteria, trait dependencies, adverse events
Introduction
Randomized controlled trials are the gold standard for generating high-quality medical evidence. An important expectation of any clinical trial is that the results are generalizable, extending to patients with the same condition and for whom the treatment is intended but who are not in the study. In other words, generalizability measures whether the results obtained by testing on a population sample are applicable to a larger population from which the sample was drawn. The need for generalizing study results to real-world populations should be addressed by research investigators and sponsors during the design of clinical trials.
Population representativeness of study populations is one of the determining factors for generalizability. The concepts generalizability and representativeness are related, but distinct. Representativeness measures the study population’s coverage of real-world patients, often with respect to multiple study traits—quantitative (e.g., lab results, age) or categorical (e.g., diagnosis, procedures). Generalizability is the portability of causal effects of a clinical intervention to a different situation.1–4 Besides population representativeness, multiple other factors affect the generalizability of a clinical study, such as variation in patient care across different clinical settings and discrepancies in conditions under which a trial is conducted.4–6 The major causes affecting generalizability are discussed in more detail by Bonell et al.6 and Kukull et al.7 In this study, we focused on measuring population representativeness.
One major factor affecting population representativeness is the study’s eligibility criteria. Restrictiveness of eligibility criteria can jeopardize the extrapolation of trial results to wider populations.8 Highly restrictive eligibility criteria can also result in studies that exclude patients who might benefit from the intervention or jeopardize patient safety by not recognizing possible postmarketing adverse intervention effects in untested populations.9–11 Conversely,loosely formed criteria can lower the effectiveness of the intervention in unselected populations. Several limitations exist with the current design of eligibility criteria, including criteria being (1) restrictive,12 (2) ambiguous,13 (3) reused with minimal modifications,14 (4) subjective,15 and (5) requiring frequent amendments owing to the lack of feasibility based on trial and error iterations.16 One of the main causes of the limitations of such designs is lack of information about the distribution of real-world patients.17
The emergence of vast electronic health records, comprehensive clinical data warehouses, and interoperable data networks have made enormous amounts of electronic patient data available. Some examples include public medical record databases, such as Multi-parameter Intelligent Monitoring in Intensive Care II (MIMIC II; https://mimic.physionet.org/), the international collaborative consortium for Observational Health Data Sciences and Informatics (OHDSI; http://www.ohdsi.org), the Clinical Data Warehouse (CDW) at Columbia University,18 and the Knowledge Program (KP) data warehouse.19 Access to these data infrastructures enable scalable analytics for phenotype modeling and population profiling, as well as for generating a low-bias approximation of the real-world population. Meanwhile, clinical trial registries and study result databases are becoming increasingly available publicly, especially through ClinicalTrials.gov, which is a registry for publically and privately funded clinical research studies conducted worldwide. Some sponsors are required by U.S. law to register their trials on ClinicalTrials.gov, with each study record consisting of the purpose, recruitment status, eligibility criteria, location, and contact information. The public’s trust in clinical research can benefit from such transparency in study information. The vast data resources mentioned above can enable evaluation and optimization of eligibility criteria design using data-driven approaches. This, in turn, can provide useful decision aids to eligibility criteria designers by determining (1) which patients should be studied and which patients should be excluded, and (2) how the study population would be representative of real-world patients and enable modification of eligibility criteria on the basis of this early feasibility feedback (before the start of recruitment).
To this end, we present a metric for quantifying a priori generalizability, or population representativeness, of an individual clinical study by considering multiple eligibility criteria simultaneously. With this metric, we aim to answer the following questions about eligibility criteria: (1) How restrictive are the overall eligibility criteria?, (2) Is there a specific eligibility criterion that lowers population representativeness?, and (3) How robust is the population representativeness to small changes in eligibility criteria? Answering these questions could optimize study generalizability within the constraints of patient safety.
On this basis, we aim to answer the vital research question, How might the population representativeness of a clinical trial (measured by our metric) be correlated with the reported adverse events associated with the trial? We hypothesize that more stringent eligibility criteria may lead to study populations with less risk of adverse events.20–24 In a clinical trial, an adverse event can be any unfavorable and unintended sign, symptom, or disease temporarily associated with the intervention, without any judgment about causality or relationship to the drug.25 Serious adverse events may refer to death, life-threatening emergencies, hospitalization, congenital or birth defects (in the case of pregnant women), and other serious medical conditions. The set of non-serious adverse events is much larger and may include, for example, tachycardia, anemia, diarrhea, decrease in urine output, and headache. Reported serious adverse events have been on the rise, as shown by data collected from the U.S. Food and Drug Administration (FDA) between 1998 and 2005.26 Moreover, it was also shown that 14% of adverse events were due to organizational factors, of which 93% were preventable.23
Background
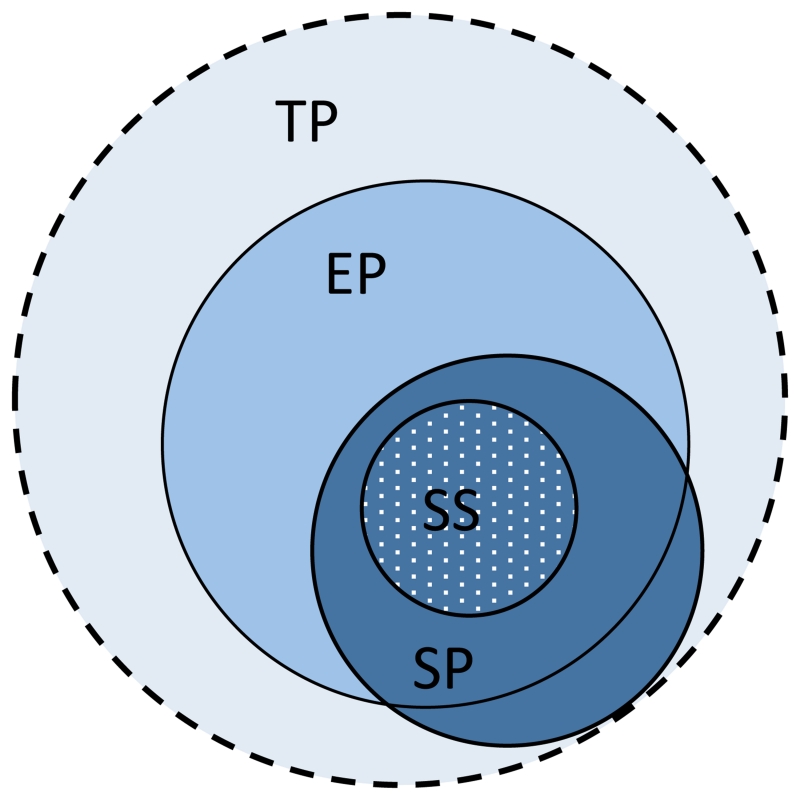
For a clinical trial, there are typically four associated populations, one of which is the target population (TP), corresponding to the entire universe of patients affected by the condition under consideration and for whom the results of the clinical trial are intended. However, since it is virtually impossible to precisely define the TP, the electronic health record (EHR) population (EP), which includes those patients who visit medical facilities to receive care, is often used as its approximation. A study population (SP) is the set of all patients who satisfy the eligibility criteria of a particular trial. Finally, the patients who actually enroll for the clinical trial constitute the study sample (SS). While part of the SP may lie outside of the EP, we assume that, during the screening for a trial, an enrolled patient’s medical data will become available electronically so that the SS will always be within the EP. A Venn diagram for these populations is shown in Figure 1. It must be noted that, in outpatient community-based trials, the EP may be a poor approximation of the TP and parts of the SS may lie outside of the EP.
Figure 1.
A schematic representation of the various populations associated with a clinical trial: target population (TP), EHR population (EP), study population (SP), and study sample (SS).
Generalizability in clinical trials is of two types: eligibility driven and sample driven. Eligibility-driven generalizability is computed prior to the trial and calculates the representativeness of the SP within the TP. Note that the SP is defined literally by the eligibility criteria. Sample-driven generalizability is calculated after a trial has been completed, using patient-level (or aggregate) data, and this computes the representativeness of the SS within the TP. As the SP subsumes the SS, the sample-driven generalizability is usually lower than eligibility-driven generalizability. Only in cases of ideal recruitment is the SS completely representative of the SP, resulting in the two generalizability calculations being equivalent. In this paper, our focus is on eligibility-driven generalizability, and unless specified otherwise, we use the term generalizability to refer to eligibility-driven generalizability throughout the rest of the paper.
Previous assessments of eligibility-driven generalizability have been based on visualization techniques and statistical tests. For example, Schoenmaker et al. used comparisons of histograms to show that TP patients are generally older than patients recruited for trials.9 Furthermore, Pressler et al. used propensity scores to estimate the generalizability bias in simulated observational data,27 and Greenhouse et al. computed generalizability bias by comparing the characteristics of SS patients to a simulated TP.28 At present, there are relatively few robust methods for quantifying generalizability during the study design process. Moreover, many of these compute sample-driven (rather than eligibility-driven) generalizability after the publication of study results.29–31
In a study by Bleeker et al,32 eligibility-driven generalizability was computed using Receiver Operating Characteristic analysis. Specifically, infants with fever were evaluated for the presence of bacterial infection by a binary classifier, and training and testing sets comprised of patients from two different hospitals in different time periods. The framework of this study is different from a clinical trial, as ground truth knowledge cannot be assumed in clinical trial situations. Weng et al. introduced the generalizability index for study traits (GIST) as a figure of merit for assessing generalizability.17 In its most general form, GIST could compute the generalizability of multiple related trials (of the same disease) at the same time and was initially presented for individual study traits, but was later extended to multiple traits by He et al.33
However, GIST had a few limitations. Firstly, all clinical traits were considered independently and the dependencies between them were not accounted for. In addition, every trait was equally weighted (i.e., all traits were assumed to be of equal significance), which is contrary to the way that traits are weighted in a clinical trial setting, where some traits may be more significant than others; this needs to be incorporated into the quantification. With these limitations in mind, we developed GIST 2.0. To differentiate between the two versions of GIST we refer to them as GIST 1.0 and GIST 2.0 (the shorthand GIST will also refer to GIST 2.0). In this paper, we discuss a pilot study aimed to illustrate the methodology of GIST 2.0 using sixteen sepsis trials (all of which reported adverse events) and follow with an analysis of the correlation between population representativeness and adverse events.
Methods
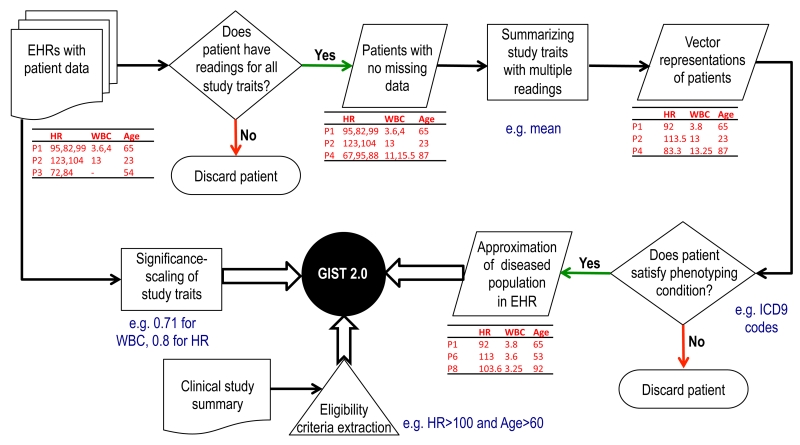
The computation of the GIST 2.0 metric has three major preprocessing steps. First, the TP is approximated using patient-level EHR data. Second, each study trait is assigned a significance, and, finally, the eligibility criteria for the trial under consideration are extracted from clinical trial summaries. The three preprocessing steps leading to the computation of the GIST 2.0 scores are shown schematically in Figure 2.
Figure 2.
A flowchart showing the various preprocessing steps to prepare the data for the computation of GIST 2.0.
Approximation of the target population
We used the EP as an approximation for the TP. Although our methodology can be applied to EHR Big Data of hundreds of millions of patients, to make it convenient for others to validate the methodology and replicate the results, we used the public data from MIMIC II for this study. MIMIC II contains structured EHRs of over 30,000 intensive care unit (ICU) patients. To illustrate the ability of the methods to handle a large number of traits, we considered sepsis trials, which usually have many traits in their eligibility criteria. Thirteen study traits, which are frequently used in sepsis trials, were chosen after consultations with a medical expert (Andrew Goldstein) and were extracted for all of the patients. These traits were bilirubin, platelet count, glucose, heart rate (HR), white blood cell (WBC) count, international normalized ratio (INR), mean arterial blood pressure (MABP), systolic blood pressure (SBP), diastolic blood pressure (DBP), respiratory rate (RR), plasma lactate, temperature (°C), and activated partial thromboplastin time (APTT). In addition, since age and gender are part of the eligibility criteria for all trials, they were added to the set of traits for GIST 2.0 calculation.
Out of all patients, 4617 had at least one value for each of the 15 traits. Owing to the large number of laboratory traits used in sepsis, we used the presence of the readings for all of the sepsis-related traits as an indicator for sepsis cases. Therefore, these 4617 patients formed our EP. We used the mean to summarize traits that had multiple readings. Identification of sepsis patients through International Classification of Diseases (ICD)-9 diagnosis codes was not accurate because of the incompleteness of data. Approximately 500 patients had no diagnosis codes and several hundred patients had incomplete sets of codes (e.g., a condition mentioned in the clinical notes was not present in the diagnosis codes list).
Significance scaling of study traits
Eligibility traits have varying clinical significance. Assigning a significance rating to each trait requires disease-specific domain knowledge. We used a simplified data prevalence-based significance rating. The significance for a trait was defined as the fraction of patients in the data set who had at least one reading for that trait.
Selection of trials and interpretation of eligibility criteria
We searched for sepsis trials in ClinicalTrials.gov that presented results and found 62 such trials. Several of these trials were actually for related but different diseases, such as bacteremia and candidemia, and were thus excluded from this study. Trials for neonatal and pediatric sepsis were also excluded. This left 16 sepsis trials that were used for further analysis. Since the number of traits in MIMIC II is limited, any additional traits in these trials (besides the abovementioned traits) were ignored.
Sepsis clinical trial eligibility criteria tend to be vague in their definition and required some clinical interpretation. The 16 trials broadly fit into three disease subcategories: sepsis, severe sepsis, and septic shock. However, the distinctions between these categories are not strictly defined.34 In the present study, we used the following definitions. The broad category of sepsis (unless defined otherwise) was interpreted as satisfying at least two of the four systemic inflammatory response syndrome (SIRS) criteria: (1) core temperature higher than 38 °C or lower than 36 °C, (2) RR greater than 20 breaths per minute, (3) WBC count greater than 12,000 per mm3 or less than 4000 per mm3, and (4) HR greater than 90 beats per minute. Severe sepsis is characterized by the SIRS criteria accompanied by signs of organ failure. In case the conditions for organ failure were not specified, we used the following conditions as indicative of organ failure:35 serum lactate greater than 2 mmol/L, platelet count less than 100,000, bilirubin greater than 2 mg/dL, or INR greater than 1.5. Septic shock requires the fulfillment of the SIRS criteria, advanced organ failure, as well as hypotension (SBP < 90 mmHg or MABP < 65 mmHg), or serum lactate levels greater 4 mmol/L, after adequate fluid resuscitation.
Brief methodology for GIST 2.0
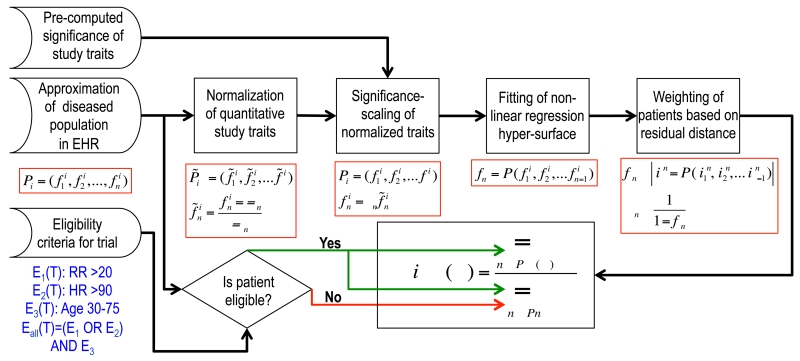
In this section we briefly describe the methods for computing a priori generalizability. The patients in the EP are denoted by P1,P2, …,PN and the traits by f1, f2, …, PN. Patients are indexed by i and traits by j. An n-dimensional vector of traits can then represent a patient, . Since each trait has a different range of values, we normalized the traits with the corresponding Z-scores (using the mean μj and standard deviation σj of a trait fj). These normalized traits were then significance scaled. We employed a nonlinear regression hyper-surface36 to model the dependencies between the normalized and significance-scaled traits, in which one of the traits was treated as the dependent variable and all others as independent (e.g, fn = F(f1, f2,… ,fn-1). Next, each patient Pi was assigned a weight wi that was inversely proportional to its residual distance from the regression hyper-surface.
For a trial T, the eligibility range (determined by eligibility criteria) for a study trait fj is denoted as Ej(T). The logical combinations of the individual eligibility criteria determined the overall eligibility criterion for T, Eall(T). Now, the multiple-trait GIST (mGIST) of T is defined as the weighted fraction of patients satisfying the eligibility criteria.
This process of computing mGIST is outlined in Figure 3. We similarly define the single-trait GIST (sGIST) for each trait by calculating the weighted fraction of patients who satisfy the eligibility criteria for that particular trait.
Figure 3.
A summary of the steps involved in computing mGIST 2.0.
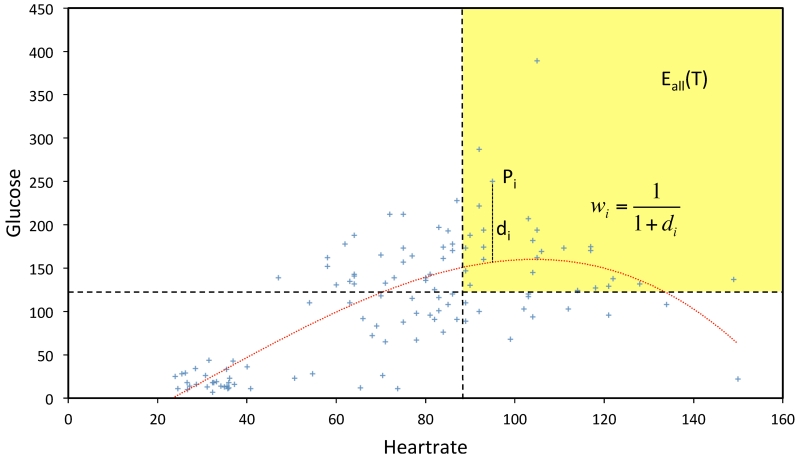
This process is demonstrated with a simple example using two traits, as shown in Figure 4. For simplicity, the normalization and the significance-scaling steps are not shown in this figure and the nonlinear regression hyper-surface was calculated directly from the observed study traits. The axes represent the heart rate and glucose measurements of 120 randomly selected patients from the EP described above. The dashed lines denote typical sepsis eligibility criteria for these two traits (HR > 90 beats per minute, glucose > 120 mg/dL). Patients in the yellow region satisfy both eligibility criteria. The dotted nonlinear curve (calculated by nonlinear regression) models the dependence between the two traits. For a patient Pi, its residual distance from this curve is di. Then the weight assigned to it is wi = 1/(1 + di).
Figure 4.
An illustration of the hyper-surface fitting and the patient weighting within the GIST 2.0 methodology using two study traits—heart rate and glucose.
Generalizability versus adverse events
We used mGIST to analyze the correlation between generalizability and the occurrence of adverse events. Trial results in ClinicalTrials.gov contain detailed information about serious and other adverse events (both grouped by cohorts). We define the total adverse events (TAEs) as the sum of serious and non-serious adverse events. For each trial, we computed the serious adverse event fraction (SAEF) and total adverse event fraction (TAEF) as the fraction of patients (from all cohorts) who suffered SAEs or any adverse events, respectively. We used these to compute the weighted correlation coefficients37 between mGIST and T/SAEs. The number of patients in each trial determined the weights for trial. The correlation coefficient measures the linear dependence between two variables and is bounded by –1 and 1, with the bounds representing perfect negative (inverse) and perfect positive (direct) correlations, respectively. For example, a strong positive correlation (with coefficient 0.71) was demonstrated between hemoglobin A1C and blood glucose by Sikaris.38
Results
The mGIST scores for the sixteen trials (in ascending order) along with their SAEF and TAEF are given in Table 1. The number of patients in the trials ranged from 5 to 745. Of the five trials with no reported adverse events, only one had more than 24 patients (NCT01947127 with 292 patients). Sepsis trials usually have complicated eligibility criteria with several logical connectors. Hence, we specify these conditions in the last column of Table 1 (without units), in which SIRS (unless specified otherwise) refers to at least two of the SIRS criteria being satisfied. Organ failures are defined as above. In addition, a comma implies a logical AND operation.
Table 1. The mGIST scores, SAEF, and TAEF for 16 sepsis trials and their eligibility criteria.
| Trial | mGIST | SAEF | TAEF | Eligibility criteria |
|---|---|---|---|---|
| NCT00979121 | 0.113 | 0.089 | 0.089 | All SIRS conditions except RR, age > 18 years |
| NCT01443494 | 0.167 | 0.000 | 0.000 | SIRS, SBP < 90 or MABP < 65, age > 18 years |
| NCT01689441 | 0.167 | 0.000 | 0.000 | SIRS, SBP < 90 or MABP < 65, age > 18 years |
| NCT00535821 | 0.210 | 0.000 | 0.000 | SIRS, lactate > 4 or SBP < 90, age > 18 years |
| NCT01150409 | 0.210 | 0.818 | 0.818 | SIRS, lactate > 4 or SBP < 90, age > 18 years |
| NCT00608322 | 0.211 | 0.306 | 0.306 | SIRS, lactate > 4 or SBP < 90, age > 14 years |
| NCT01947127 | 0.217 | 0.000 | 0.000 | SIRS, MABP > 70, SBP > 90, age 18–65 years |
| NCT00633477 | 0.289 | 0.235 | 0.824 | At least 3 of the SIRS conditions, age > 18 years |
| NCT01434121 | 0.315 | 0.000 | 0.000 | SIRS, lactate > 2.5 or platelet < 80,000 or MABP < 60, age > 18 years |
| NCT00386425 | 0.380 | 0.167 | 0.366 | SIRS, 2 or more organ failures, age > 18 years |
| NCT01027897 | 0.604 | 0.267 | 0.267 | SIRS, age 18–90 years |
| NCT01144624 | 0.609 | 0.300 | 1.000 | SIRS, age > 20 years |
| NCT01145560 | 0.611 | 0.289 | 0.289 | SIRS, age > 18 years |
| NCT01739361 | 0.611 | 0.068 | 0.068 | SIRS, age > 18 years |
| NCT00464204 | 0.611 | 0.495 | 0.989 | SIRS, age > 18 years |
| NCT00279214 | 0.612 | 0.163 | 0.256 | SIRS, age > 18 years |
Abbreviations: mGIST, multiple-trait generalizability index for study traits; SAEF, serious adverse event fraction; TAEF, total adverse event fraction; SBP, systolic blood pressure; MABP, mean arterial blood pressure; SIRS, systemic inflammatory response syndrome.
The weighted correlation coefficient for mGIST and the SAEF was 0.709 and was higher (0.0825) for mGIST and the TAEF. Both of these coefficients were statistically significant at the 0.01 level (P = 0.0021 and < 0.0001, respectively), demonstrating a positive dependence between mGIST and occurrence of adverse events. The trials that have more restrictive eligibility criteria (thus lowering their generalizability) usually have fewer cases of adverse events. However, these correlations should be interpreted with caution because of the reasons discussed in the next section.
Although the logical combinations of eligibility criteria are complicated (and different) for sepsis trials, the criteria for the individual traits received almost identical sGIST scores. The sGIST scores for all eligibility traits in all 16 trials are shown in Table 2 and the main points are summarized here. For the four SIRS criteria, the sGIST values were 0.372 for temperature, 0.535 for lactate, 0.435 for RR, and 0.512 for HR. For SBP less than 90 mmHg, the sGIST was 0.159, while for MABP less than 65 mmHg, it was 0.191 (0.115 for less than 60 mmHg). Serum lactate had three different eligibility criteria: > 2, > 2.5 mmol/L, and > 4 mmol/L. The sGISTs for these cases were 0.483, 0.370, and 0.189, respectively. For the organ failure criteria, the sGISTs were: 0.187 for bilirubin, 0.390 for INR, and 0.159 for platelet count (0.107 when the criterion was < 80,000). The sGIST for age greater than 18 years was 0.999 and for the 18–65 year range, the score was 0.501.
Table 2. The sGIST scores for each study trait in 16 sepsis trials and the trials’ mGIST scores.
| Bilirubin | Lactate | Temp. | INR | WBC | Platelet | MABP | SBP | DBP | RR | HR | Age | mGIST | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCT00979121 | 0.370 | 0.534 | 0.512 | 0.999 | 0.113 | ||||||||
| NCT01443494 | 0.372 | 0.191 | 0.160 | 0.435 | 0.513 | 0.999 | 0.167 | ||||||
| NCT01689441 | 0.371 | 0.190 | 0.159 | 0.435 | 0.512 | 0.999 | 0.167 | ||||||
| NCT00535821 | 0.189 | 0.371 | 0.159 | 0.435 | 0.512 | 0.999 | 0.210 | ||||||
| NCT01150409 | 0.190 | 0.372 | 0.160 | 0.436 | 0.513 | 0.999 | 0.210 | ||||||
| NCT00608322 | 0.189 | 0.372 | 0.160 | 0.436 | 0.513 | 1.000 | 0.211 | ||||||
| NCT01947127 | 0.372 | 0.721 | 0.849 | 0.435 | 0.513 | 0.501 | 0.217 | ||||||
| NCT00633477 | 0.372 | 0.436 | 0.513 | 0.999 | 0.289 | ||||||||
| NCT01434121 | 0.370 | 0.372 | 0.535 | 0.107 | 0.115 | 0.436 | 0.513 | 0.999 | 0.315 | ||||
| NCT00386425 | 0.187 | 0.483 | 0.371 | 0.159 | 0.535 | 0.159 | 0.191 | 0.160 | 0.530 | 0.435 | 0.513 | 0.999 | 0.380 |
| NCT01027897 | 0.372 | 0.436 | 0.513 | 0.986 | 0.604 | ||||||||
| NCT01144624 | 0.372 | 0.436 | 0.513 | 0.996 | 0.609 | ||||||||
| NCT01145560 | 0.371 | 0.435 | 0.513 | 0.999 | 0.611 | ||||||||
| NCT01739361 | 0.372 | 0.435 | 0.513 | 0.999 | 0.611 | ||||||||
| NCT00464204 | 0.371 | 0.435 | 0.512 | 0.999 | 0.611 | ||||||||
| NCT00279214 | 0.372 | 0.436 | 0.513 | 0.999 | 0.612 |
Abbreviations: Temp., temperature; INR, international normalized ratio; WBC, white blood cell; MABP, mean arterial blood pressure; SBP, systolic blood pressure; DBP, diastolic blood pressure; RR, respiratory rate; HR, heart rate; mGIST, multiple-trait generalizability index for study traits.
Discussion
While the interaction between mGIST and adverse events is interesting, their high and significant correlation should be treated with caution—although population representativeness is desirable, it cannot come at the cost of patient safety. Most trials are designed to minimize adverse events. Hence, the tradeoff between population representativeness and adverse events is challenging. Due to a limited number of relevant trials with results, our analysis considered only sixteen trials. As mentioned earlier, both the SAEF and TAEF are aggregated over all cohorts, and there may be significant differences between the various cohorts. Moreover, the adverse event could be independent of the clinical trial intervention. Despite these uncertainties, the significant positive correlation indicates that trials that have stricter eligibility criteria are less likely to have serious adverse events. This is in agreement with previous studies20,21 that have concluded that proper screening can lower the risk of adverse events, which also validates the clinical meaningfulness of mGIST.
The present study is only a stepping stone toward the leverage of Big Data resources in clinical trial generalizability assessment. The MIMIC II data set of approximately 30,000 patients is relatively small as compared to typical clinical data warehouses (e.g., the Columbia University CDW consists of 4.5 million patients’ medical records) or large international networks (e.g., OHDSI has approximately 680 million patient records as of January 2016). However, the methods presented here can be easily extended to larger data sets. The algorithm for calculating GIST 2.0 scores (though not optimized professionally) is computationally robust and can compute the fitting hyper-surface (in 15 dimensions) in less than a second. Although the addition of patients and traits will amplify the computation, it is expected to remain within manageable limits.
In contrast to GIST 1.0, which computes the multiple-trait generalizability by simply aggregating the single-trait generalizabilities, GIST 2.0 explicitly models the inter-trait dependencies. The weighing scheme for the patients in GIST 2.0 minimizes the effect of outliers. The nonlinear regression model for trait dependencies is capable of handling both categorical and numeric traits with different significance. In this study, since the analysis was limited to sepsis trials, the ability of GIST 2.0 to handle categorical variables was not evident in the results. For a full evaluation, we will expand this analysis to multiple diseases. However, despite this promise, other challenges need to be overcome. It must be noted that, by reusing EHRs for clinical research, we are using data for a purpose different from the purpose for which they were acquired, potentially causing major hurdles, particularly sampling bias and data incompleteness.39
The mGIST metric has several of the desired mathematical properties. If no patients from the TP satisfy the eligibility criteria of a trial, the mGIST of the trial is zero. Similarly, if all patients of the TP are eligible for the trial, then its mGIST is one. One of the most meaningful properties is the monotonicity property (i.e., if the eligibility criteria of a trial T1 subsumes the criteria of another trial T2, then mGIST(T1) is greater than mGIST(T2). The distributive law also holds, in that when the SP (defined by eligibility criteria) of a trial is partitioned into two or more components, the sum of the mGISTs of the components is equal to the mGIST of the trial. For example, if a trial open to both genders is restricted to a single gender, then its mGIST is halved (assuming an equal distribution of genders and no third gender). As a follow-up of the monotonicity property, eligibility criteria connected by the logical AND lower the mGIST, while those connected by OR elevate the mGIST.
Our study has certain limitations; namely, it is debatable how well the EP can approximate the TP. For example, it has previously been shown that the EP is younger9 (owing to a higher number of medical records of recent patients) and sicker.40 Our methodology for defining the EP was based on the presence of 15 sepsis-related traits; however, such a selection may introduce biases within the EP. Imputation and interpolation are possible ways for dealing with patients with missing traits. Further, this method for identifying sepsis patients may include some patients in the EP who do not have sepsis. Therefore, in the future we plan to use better data sets, such as OHDSI, which encompass a far greater variety of patients. Another way to enrich the data is the use of claims data along with EHRs.41 The eligibility-driven generalizability, as defined here, depends only on the traits that are part of the eligibility criteria and not on thousands of other latent traits, many of which may be clinically relevant. This can lead to the SP being very different from the EP and TP.
Conclusions
We have applied a quantitative metric GIST 2.0 for evaluating the population representativeness of a clinical trial and correlated it with study adverse events. Initial results point toward a robust metric with all of the desired mathematical properties.
Acknowledgments
This study is supported by National Library of Medicine Grant R01LM009886 (PI: Weng). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Fernandez-Hermida JR, Calafat A, Becoña E, Tsertsvadze A, Foxcroft DR. Assessment of generalizability, applicability and predictability (GAP) for evaluating external validity in studies of universal family-based prevention of alcohol misuse in young people: systematic methodological review of randomized controlled trials. Addiction. 2012;107:1570–1579. doi: 10.1111/j.1360-0443.2012.03867.x. [DOI] [PubMed] [Google Scholar]
- 2.Neely AS, Backman L. Effects of Multifactorial Memory Training in Old Age: Generalizability across Tasks and Individuals. Journals Gerontol. Ser. B Psychol. Sci. Soc. Sci. 1995;50B:P134–P140. doi: 10.1093/geronb/50b.3.p134. [DOI] [PubMed] [Google Scholar]
- 3.Weiss NS. Generalizability of cancer clinical trial results. Cancer. 2007;109:341–341. doi: 10.1002/cncr.22390. [DOI] [PubMed] [Google Scholar]
- 4.Pearl J. Generalizing Experimental Findings. J. Causal Inference. 2015;3:259–266. [Google Scholar]
- 5.Slack MK, Draugalis JR. Establishing the internal and external validity of experimental studies. Am. J. Health. Syst. Pharm. 2001;58:2173–81. quiz 2182-3. [PubMed] [Google Scholar]
- 6.Bonell C, Oakley A, Hargreaves J, Strange V, Rees R, Chinnock P, Siegfried N, Clarke M, Moher D, Schulz K, Altman D, Kraft J, Mezoff J, Sogolow E, Spink NM, Thomas P, Oakley A, Strange V, Bonell C, Allen E, Stephenson J, Bartlett C, Doyal L, Ebrahim S, Davey P, Backmann M, Egger M, Rees R, Harden A, Thomas J, Oliver S, Kavanagh J, Burchett H, Dilley J, Woods W, Sabatino J, Lihatsh T, Adler B, Casey S, Elford J, Bolding G, Sherr L, Elford J, Sherr L, Bolding G, Serle F, Maguire M, Elford J, Sherr L, Bolding G, Maguire M, Serle F, Flowers P, Hart G, Williamson L, Frankis J, Der G, Flowers P, Hart G, Gold R, Rosenthal D, Imrie J, Stephenson J, Cowan F, Wanigaratne S, Billington A, Copas A, Picciano J, Roffman R, Kalichman S, Rutledge S, Berghuis J, Rosser B, Bochting B, Rugg D, Robinson B, Ross M, Bauer G, Shepherd J, Weare K, Turner G, Shepherd J, Turner G, Weare K, Campbell M, Fitzpatrick R, Haines A, Kinmouth A, Sandercock P, Spiegelhalter D. Assessment of generalisability in trials of health interventions: suggested framework and systematic review. BMJ. 2006;333:346–9. doi: 10.1136/bmj.333.7563.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kukull WA, Ganguli M. Generalizability The trees, the forest, and the low-hanging fruit. Neurology. 2012;78:1886–1891. doi: 10.1212/WNL.0b013e318258f812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bijker N, Peterse JL, Fentiman IS, Julien JP, Hart AA, Avril A, Cataliotti L, Rutgers EJT. Effects of patient selection on the applicability of results from a randomised clinical trial (EORTC 10853) investigating breast-conserving therapy for DCIS. Br. J. Cancer. 2003;87:615–620. doi: 10.1038/sj.bjc.6600514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenmaker N, Van Gool WA. The age gap between patients in clinical studies and in the general population: a pitfall for dementia research. Lancet Neurol. 2004;3:627–630. doi: 10.1016/S1474-4422(04)00884-1. [DOI] [PubMed] [Google Scholar]
- 10.Masoudi FA, Havranek EP, Wolfe P, Gross CP, Rathore SS, Steiner JF, Ordin DL, Krumholz HM. Most hospitalized older persons do not meet the enrollment criteria for clinical trials in heart failure. Am. Heart J. 2003;146:250–7. doi: 10.1016/S0002-8703(03)00189-3. [DOI] [PubMed] [Google Scholar]
- 11.Ma H, Weng C. Identification of questionable exclusion criteria in mental disorder clinical trials using a medical encyclopedia. Pac Symp Biocomput. 2016;21:219–230. [PMC free article] [PubMed] [Google Scholar]
- 12.Musen MA, Rohn JA, Fagan LM, Shortliffe EH. Knowledge engineering for a clinical trial advice system: Uncovering errors in protocol specification. Bull. Cancer. 1987;74:291–296. [PubMed] [Google Scholar]
- 13.Ross J, Tu SW, S C, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl. Sci. Proc. 2010:46–50. [PMC free article] [PubMed] [Google Scholar]
- 14.Hao T, Rusanov A, Boland MR, Weng C. Clustering clinical trials with similar eligibility criteria features. J. Biomed. Inform. 2014;52:112–120. doi: 10.1016/j.jbi.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin DL, Gennari J, Musen MA. Knowledge representation and tool support for critiquing clinical trial protocols. Proc. AMIA Annu. Symp. 2000:724–728. [PMC free article] [PubMed] [Google Scholar]
- 16.Weng C. Optimizing Clinical Research Participant Selection with Informatics. Trends Pharmacol. Sci. 2015;36:706–709. doi: 10.1016/j.tips.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weng C, Li Y, Ryan P, Zhang Y, Liu F, Gao J, Bigger J, Hripcsak G. A Distribution-based Method for Assessing The Differences between Clinical Trial Target Populations and Patient Populations in Electronic Health Records. Appl. Clin. Inform. 2014;5:463–479. doi: 10.4338/ACI-2013-12-RA-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson SB. Generic data modeling for clinical repositories. J. Am. Med. Inform. Assoc. 1996;3:328–39. doi: 10.1136/jamia.1996.97035024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzan I, Speck M, Dopler C, Urchek J, Bielawski K, Dunphy C, Jehi L, Bae C, Parchman A. The Knowledge Program: an innovative, comprehensive electronic data capture system and warehouse. AMIA Annu. Symp. Proc. 2011;2011:683–92. [PMC free article] [PubMed] [Google Scholar]
- 20.Tai M, Vu L, Bostwick JR. Antidepressant Treatment Concerns and Recommendations for Avoiding Adverse Events. US Pharm [Google Scholar]
- 21.Pretorius RW, Gataric G, Swedlund SK, Miller JR. Reducing the Risk of Adverse Drug Events in Older Adults. Am Fam Physician. 2013;87:331–336. [PubMed] [Google Scholar]
- 22.Karson AS, Bates DW. Screening for adverse events. J. Eval. Clin. Pract. 1999;5:23–32. doi: 10.1046/j.1365-2753.1999.00158.x. [DOI] [PubMed] [Google Scholar]
- 23.Smits M, Zegers M, Groenewegen PP, Timmermans DRM, Zwaan L, van der Wal G, Wagner C. Exploring the causes of adverse events in hospitals and potential prevention strategies. BMJ Qual. Saf. 2010;19:e5–e5. doi: 10.1136/qshc.2008.030726. [DOI] [PubMed] [Google Scholar]
- 24.Ory M, Resnick B, Jordan PJ, Coday M, Riebe D, Ewing Garber C, Pruitt L, Bazzarre T. Screening, safety, and adverse events in physical activity interventions: collaborative experiences from the behavior change consortium. Ann. Behav. Med. 2005;29 Suppl:20–8. doi: 10.1207/s15324796abm2902s_5. [DOI] [PubMed] [Google Scholar]
- 25.International Council for Harmonisation Guidance for Industry - Good Clinical Practice: Consolidated Guidance. 1996;20857:301–827. [Google Scholar]
- 26.Moore TJ, Cohen MR, Furberg CD. Serious adverse drug events reported to the Food and Drug Administration, 1998-2005. Arch. Intern. Med. 2007;167:1752–1759. doi: 10.1001/archinte.167.16.1752. [DOI] [PubMed] [Google Scholar]
- 27.Pressler TR, Kaizar EE. The use of propensity scores and observational data to estimate randomized controlled trial generalizability bias. Stat. Med. 2013;32:3552–3568. doi: 10.1002/sim.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenhouse JB, Kaizar EE, Kelleher K, Seltman H, Gardner W. Generalizing from clinical trial data: A case study. The risk of suicidality among pediatric antidepressant users. Stat. Med. 2008;27:1801–1813. doi: 10.1002/sim.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchanan A, Hudgens M, Cole S, Mollan K, Sax P, Daar E, Adimora A, Eron J, Mugavero M. Generalizing Evidence from Randomized Trials using Inverse Probability of Sampling Weights. Univ. North Carolina Chapel Hill Dep. Biostat. Tech. Rep. Ser. 2015 doi: 10.1111/rssa.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang TS, Hellkamp AS, Patel CB, Ezekowitz JA, Fonarow GC, Hernandez AF. Representativeness of RELAX-AHF clinical trial population in acute heart failure. Circ. Cardiovasc. Qual. Outcomes. 2014;7:259–268. doi: 10.1161/CIRCOUTCOMES.113.000418. [DOI] [PubMed] [Google Scholar]
- 31.Susukida R, Crum RM, Stuart EA, Ebnesajjad C, Mojtabai R. Assessing Sample Representativeness in Randomized Control Trials: Application to the National Institute of Drug Abuse Clinical Trials Network. Addiction. 2016 doi: 10.1111/add.13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bleeker SE, Moll HA, Steyerberg EW, Donders ART, Derksen-Lubsen G, Grobbee DE, Moons KGM. External validation is necessary in prediction research: a clinical example. J. Clin. Epidemiol. 2003;56:826–32. doi: 10.1016/s0895-4356(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 33.He Z, Ryan P, Hoxha J, Wang S, Carini S, Sim I, Weng C. Multivariate analysis of the population representativeness of related clinical studies. J. Biomed. Inform. 2016;60:66–76. doi: 10.1016/j.jbi.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marik PE, Lipman J. The definition of septic shock: implications for treatment. Crit. Care Resusc. 2007;9:101–103. [PubMed] [Google Scholar]
- 35.Stony Brook Medicine - Severe Sepsis/Septic Shock: Recognition and Treatment Protocols. 2013. [Google Scholar]
- 36.Hypersurface at < http://mathworld.wolfram.com/Hypersurface.html>.
- 37.Weighted Correlation Coefficients. at < https://en.wikipedia.org/wiki/Pearson_product-moment_correlation_coefficient#Weighted_correlation_coefficient>.
- 38.Sikaris K. The correlation of hemoglobin A1c to blood glucose. J. Diabetes Sci. Technol. 2009;3:429–38. doi: 10.1177/193229680900300305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiskopf NG, Hripcsak G, Swaminathan S, Weng C. Defining and measuring completeness of electronic health records for secondary use. J. Biomed. Inform. 2013;46:830–836. doi: 10.1016/j.jbi.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiskopf NG, Rusanov A, Weng C. Sick patients have more data: the non-random completeness of electronic health records. AMIA Annu. Symp. Proc. 2013:1472–1477. [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson J. The benefit of using both claims data and electronic medical record data in health care analysis. Optum Insight. 2012:1–4. [Google Scholar]