Abstract
The nematode Caenorhabditis elegans is well established as a system for characterization and discovery of molecular mechanisms mediating microbe-specific inducible innate immune responses to human pathogens. Coxiella burnetii is an obligate intracellular bacterium that causes a flu-like syndrome in humans (Q fever), as well as abortions in domesticated livestock, worldwide. Initially, when wild type C. elegans (N2 strain) was exposed to mCherry-expressing C. burnetii (CCB) a number of overt pathological manifestations resulted, including intestinal distension, deformed anal region and a decreased lifespan. However, nematodes fed autoclave-killed CCB did not exhibit these symptoms. Although vertebrates detect C. burnetii via TLRs, pathologies in tol-1(−) mutant nematodes were indistinguishable from N2, and indicate nematodes do not employ this orthologue for detection of C. burnetii. sek-1(−) MAP kinase mutant nematodes succumbed to infection faster, suggesting that this signaling pathway plays a role in immune activation, as previously shown for orthologues in vertebrates during a C. burnetii infection. C. elegans daf-2(−) mutants are hyper-immune and exhibited significantly reduced pathological consequences during challenge. Collectively, these results demonstrate the utility of C. elegans for studying the innate immune response against C. burnetii and could lead to discovery of novel methods for prevention and treatment of disease in humans and livestock.
Keywords: Coxiella burnetii, Caenorhabditis elegans, human pathogenic bacteria, immunity, Q fever
Introduction
Coxiella burnetii is a highly infectious, obligate intracellular γ-proteobacterium that causes Q fever in humans.1 Q fever is a zoonosis maintained in nature between hematophagous ticks,2 and a wide variety of domesticated (goats, sheep, cattle) and wild animals (e.g. kangaroos, foxes, cats, dogs).3–7 It is generally accepted that domesticated ruminants are primary reservoirs of C. burnetii worldwide. Coxiellosis is the term used to describe pathologies associated with a C. burnetii infection of livestock and can include colonization of the placenta, stillbirth and premature abortion.8 Infected animals shed bacteria through contaminated milk, placenta, urine, amniotic fluid and feces.
Humans living in close proximity to infected herds and those who care for livestock (e.g. ranchers, veterinarians, abattoir employees) are at highest risk of contracting Q fever.1,9,10 It is generally accepted that inhalation of contaminated aerosols containing the environmentally resistant spore-like morphotype of C. burnetii is the primary route of human infection.11 This spore-like morphotype, or small-cell variant (SCV), is highly resistant to environmental stress (heat, desiccation, UV light). Environmental persistence, combined with its characteristic low infectious dose, aerosol-borne transmission to humans and past use as a bio-weapon resulted in classification of C. burnetii as a CDC Category B select agent.
Reports describing Q fever outbreaks throughout the world have been steadily increasing.12–18 An epidemic recently occurred in the Netherlands over a 4-yr period (2007–2011), wherein >4000 cases of acute Q fever were reported, and approximately 50% of patients were hospitalized.19,20 In an attempt to control the dramatic rise in cases, 957 pregnant sheep and goats in 13 herds positive for Coxiella were culled and remaining animals were vaccinated.21 This event demonstrates the significant impact of C. burnetii on the health of domesticated livestock and the human food supply.
Following inhalation, SCVs transition to the metabolically active morphotype [large cell variants (LCVs)], which initially occurs in phagolysosome-like acidic compartments of alveolar macrophages (referred to as parasitophorous vacuoles).22 Over time, non-inflammatory intra-alveolar lesions develop, as well as subsequent acute and potentially chronic Q-fever pathologies. Acute manifestations in humans include a flu-like pneumonia that typically occurs within 2–3 wk following exposure to C. burnetii, often accompanied by a dry cough, fever, chills, myalgia, and malaise.1 Although most cases are self-limiting, chronic manifestations are reported in about 5% of cases, including culture-negative endocarditis, chronic hepatitis and chronic fatigue syndrome, and these patients are often prescribed years of antimicrobial therapy.23–26
C. burnetii infection has been studied in a wide variety of systems, including cultured mammalian and tick cells, as well as vertebrate and invertebrate animals.27,28 Both innate and adaptive immune responses play roles in controlling Coxiella infection of vertebrates.29 TLR and NOD play significant roles in the initial detection of Coxiella.30–32 In humans, IFN-γ-orchestrated systemic cell-mediated mechanisms lead to granuloma formation and potential resolution of acute infection. In contrast, a faulty cell-mediated immune response leads to defective granuloma formation, overproduction of IL-10 and IFN-γ, and decreased production of IL-2, resulting in subsequent chronic manifestations.29,33
Free-living nematodes such as Caenorhabditis elegans are non-segmented invertebrate roundworms that inhabit a variety of aquatic and terrestrial environments throughout the world and ingest bacteria, fungi and other organic material to develop and generate progeny.34 Free-living nematodes are capable of differentiating food from pathogens by olfactory mechanisms, and this avoidance behavior is the first innate immune response for C. elegans.35,36 The second innate immune mechanisms are physical; provided by a strong exoskeleton, and mechanical via a pharangeal grinder capable of destroying most microbes ingested upon feeding.35 The third defense mechanism consists of a complex inducible innate immune system that regulates generation of antimicrobial effectors in a pathogen-specific manner.35,37
Invertebrates of the phylum Nematoda, such as C. elegans, do not possess an apparent acquired immune response, circulating immune cells, homologues of vertebrate cytokines, NF-κB transcription factor or other components of the TLR signaling pathway such as MYD88.38 However, C. elegans does possess a number of innate immune components that are evolutionarily conserved with vertebrates, including a single TLR, p38 MAPK, FOXO and β-catenin,38 as well as TFEB-family transcription factor HLH-30, mediating response to infection.39
Over 40 human pathogens have been analyzed in the genetically tractable C. elegans infection model.40–43 Moreover, unique and specific pathologies and corresponding immune responses have been described in C. elegans. One of the most interesting models resembles the flea proventricular colonization by Yersinia pestis,44 wherein the bacterium forms a biofilm on the mouth of C. elegans.45 The overall goal of this study was to determine the utility of C. elegans as a tool for analysis of the innate immune response against C. burnetii by studying the nematode’s avoidance, physical/mechanical and inducible innate immune responses upon exposure to the pathogen.
Materials and methods
Cultivation and manipulation of bacteria
C. burnetti (Nine-Mile phase II; strain RSA 439) was cultured in acidified citrate cysteine medium-2 (ACCM-2), as previously described.46 Briefly, ACCM-2 broth in sterile, vented-cap polycarbonate flasks was inoculated with C. burnetii to a final concentration of 1 × 106 bacteria/ml. Flasks were incubated in a tri-gas incubator (Model NU-4950; Nuaire, Plymouth, MN, USA) maintained at 2.5% oxygen, 5% carbon dioxide, 92.5% nitrogen and 88% relative humidity at 37°C. Cultures were incubated for 7–10 d with gentle shaking, and bacteria collected by centrifugation (16,000 g, 15 min, 4°C) were prepared for nematode infection experiments (below). ACCM-2 was supplemented with chloramphenicol (5 μg/ml final concentration) when required. Transformation of C. burnetii with pKM244 (a Coxiella shuttle vector derived from pKM230,47 and kindly provided by Dr. James E. Samuel) was accomplished using published electroporation methods.48–50 Clones were isolated by limiting dilution growth in ACCM-2. The resulting, transformed mCherry-expressing C. burnetii strain was termed CCB. E. coli (strain OP50) was cultured overnight (~16 h) in lysogeny broth (LB) at 37°C with gentle shaking. Cells were collected by centrifugation (10,000 g, 10 min, 4°C). E. coli OP50 was transformed with pKM244 by standard electroporation methods and LB supplemented with chloramphenicol (20 μg/ml final concentration) when culturing. The resulting pKM244-transformed strain was termed mCherry E. coli (CEC).
Cultivation and manipulation of C. elegans
C. elegans wild type (N2 Bristol) and mutant [tol-1(nr2033), sek-1(km4), and daf-2(e1370)] strains were obtained from the Caenorhabditis Genetics Center, University of Minnesota. C. elegans was cultured on the surface of agar-solidified new nematode growth media (NNGM), as previously described.51 Briefly, NNGM base components (0.55 g/l Tris-HCl, 0.24 g/l Tris-base, 4.6 g/l tryptone, 2.0 g/l NaCl and 20 g/l agar) were combined, autoclaved, cooled to 65°C and supplemented with cholesterol (8 μg/ml final concentration) before pouring plates. NNGM plates were supplemented with chloramphenicol (final concentration of 20 μg/ml) when experiments included bacteria transformed with pKM244. C. elegans was routinely cultured at 25°C, although in experiments utilizing temperature-sensitive, loss-of-function allele daf-2(e1370), nematodes were cultured at 15°C and survival analyses were done at 20°C in parallel with N2.
Cultured bacteria were prepared for routine nematode feeding and infection experiments by washing cells twice using the centrifugation parameters above, in M9 buffer (3.0 g/l KH2PO4, 3.1 g/l Na2HPO4, 5.0 g/l NaCl and 0.25 g/l MgSO4; no pH adjustment; filter sterilized), microscopically enumerating and re-suspending in M9 to a final concentration of 1 × 109–1 × 1010 cells/ml. Volumes (100 μl or 300 μl) of bacterial suspensions were then spread onto the surface of NNGM plates (60 - or 100-mm Petri dishes, respectively), and allowed to dry. For experiments with dead bacteria, cell suspensions were autoclaved (20 min, liquid cycle) just prior to being spread onto plates.
C. elegans was synchronized by treatment with bleach as previously described,51 prior to C. burnetii challenges. Embryos were re-suspended in M9 (1–10 ml) and incubated with gentle shaking until the majority hatched (~24 h at 25°C). L1-stage larvae were then seeded onto plates containing UV-inactivated E. coli OP50 and cultured until the synchronized population of nematodes reached L4 stage (~40 h at 25°C) for subsequent use in infection experiments. UV-inactivated E. coli OP50 was used to prevent carry-over of live E. coli onto experimental C. burnetii plates. This was accomplished by exposing plates prepared with live E. coli to UV germicidal irradiation in a GS Gene Linker chamber (BioRad Inc., Hercules, CA, USA) for 90 s at a distance of 15 cm from the UV light source. The effectiveness of the treatment was confirmed by the lack of colonies following overnight outgrowth.
Pathogen avoidance assay
CEC and CCB were cultured and washed in M9 as above and NNGM plates containing four small lawns of bacteria were prepared, including: (i) live CCB, (ii) autoclave-killed CCB, (iii) live CEC and (iv) autoclave-killed CEC. C. elegans was cultured and synchronized as above. Twenty-five L1-stage or L4-stage larvae were separately transferred to the center of the plate, equidistant from the four bacterial lawns and allowed to migrate to the preferred food source(s). Each analysis was performed in triplicate. After 24 h at 25°C, the location of each nematode was recorded as being either on one of the four lawn types or outside any of them.
Fluorescence and light microscopy of nematodes
Routine analysis of C. elegans by fluorescence/light microscopy was accomplished by transferring nematodes to glass slides with M9 (10 μl) containing levamisole (250 μM), a nematode paralytic agent that enables better image capture. Nematodes were examined and imaged using a phase contrast microscope (BX51; Olympus, Center Valley, PA, USA) equipped with a fluorescence illuminator (X-Cite 120Q; Excelitas Technologies, Waltham, MA, USA) and a cooled digital color camera (DP72; Olympus) with accompanying acquisition software (DP2-BSW; Olympus).
Deformed anal region (Dar) morphology was recorded by mounting nematodes on a 5% agarose pad as previously described,52 and images were acquired using a Zeiss Axioscop with a 63× Plan-Apochromat NA 1.4 objective with differential interference optics (Zeiss, Thornwood, NY, USA) equipped with a cooled digital AxioCam MRm camera and Zen Blue acquisition software (Zeiss).
Nematodes in individual wells of liquid-based C. burnetii persistence assay plates were directly analyzed and imaged without levamisole by using an inverted fluorescence/light microscope (CKX41; Olympus) equipped with a fluorescence illuminator (U-RFL-T; Olympus) and imaged using the same camera (DP72) and acquisition software (DP2-BSW).
Transmission electron microscopic analysis of infection
In general, preparation of C. burnetii-infected C. elegans for transmission electron microscopic (TEM) analyses was accomplished as previously described.53 Briefly, L4 C. elegans was exposed to live CCB on the surface of NNGM plates for 6 d, washed three times in M9 and transferred to Karnovsky’s fixative [0.1 M phosphate buffer (PB) containing paraformaldehyde (4%, w/v) and glutaraldehyde (2.5%, w/v)] and incubated 16 h at 4°C. PB (0.2 M) was prepared by combining equal volumes of 0.2 M NaH2PO4 and 0.2 M Na2HPO4. Fixed nematodes were stained for 90 min in 0.1 M PB containing 1.0% OsO4 and 0.5% KFe(CN)6, and subsequently washed three times for 10 min, in 0.1 M PB and rinsed in distilled water. Stained worms were embedded in 2% agarose blocks and dehydrated with a graded ethanol series prior to being embedded in 100% propylene oxide resin, then transverse cross-sectioned. Sections were mounted on TEM grids and imaged with a Hitachi H-7100 transmission electron microscope in the University of Montana EMtrix Facility (Missoula, MT, USA).
Nematode longevity assays
The lifespan of C. elegans was analyzed by the presence and survival of nematodes while being constantly exposed to relatively high concentrations of both live and dead CCB or E. coli. Initially, we chemically synchronized and hatched L1 C. elegans populations that were cultivated to the L4 stage on plates containing UV-treated E. coli OP50 before starting longevity analyses. Thirty L4 nematodes were individually picked to NNGM plates containing: (i) live C. burnetii, (ii) dead C. burnetii, (iii) live E. coli or (iv) dead E. coli. Each preparation was tested in quadruplicate using four separate sets of 30 nematodes. On alternate days, surviving nematodes were transferred to fresh NNGM plates to ensure constant high-level exposure to the bacterial preparations, as well as to keep test nematodes separated from their fast-growing progeny. Experiments were conducted in parallel with control E. coli OP50 at the same temperature.
Nematodes were considered dead when they failed to respond to touch, were non-motile or if pharyngeal pumping was no longer observed. Nematodes that died as a result of crawling off the plate were censored from analysis. For each assay, nematode survival was calculated using the Kaplan–Meier method and survival differences were tested using a log-rank test (Graph Pad Prism software, version 7.0 a; GraphPad Inc., La Jolla, CA, USA) for determination of statistical significance of survival comparisons and generation of median survival times. The median survival time is the number of days required for 50% of nematodes in a given experiment to die. Dar phenotypes were scored over a 20-d time course as normal (none visible), weak (Dar-1) moderate (Dar-2) or severe (Dar-3) (see also Figure 6).
Results
Live C. burnetii exposure results in a number of pathological consequences to C. elegans
To explore the utility of C. elegans as a model system for studying C. burnetii pathogenesis, we initially exposed non-synchronized, wild type nematode populations to either live CEC or CCB as lawns on the surface of NNGM and observed interactions using stereo and fluorescence microscopy. From this work, it was apparent that live CCB exposure led to developmentally related pathological manifestations when compared with live CEC. The majority of young larvae (L1–L2) exposed to CCB exhibited decreased locomotion, stunted growth and died within 1 d (data not shown), whereas intact CCB appeared to accumulate in the intestinal tract of fourth-stage larvae (L4) and young adults, and consistently led to overt pathologies over time. Further analysis of L1–L2 stage nematodes was not performed in this study as young nematodes succumbed to Coxiella exposure prior to exhibiting other pathological manifestations necessary for scoring relative pathogenicity.
C. elegans does not recognize E. coli OP50 as a pathogen and avoids C. burnetii
Soil-dwelling nematodes, such as C. elegans, ingest bacteria, fungi and other organic material through the activity of a pharyngeal pump located in the anterior portion of the gut.54 The first innate defense of C. elegans is the ability to recognize and avoid noxious microorganisms and substances, as well as to migrate toward useful food sources.55 This pathogen avoidance behavior has been attributed to the ability to detect specifically products generated by bacteria (e.g. homoserine lactone autoinducers, surfactants, oxygen concentration), and is mediated by G protein-coupled, insulin-like and neuronal serotonin-signaling mechanisms.56
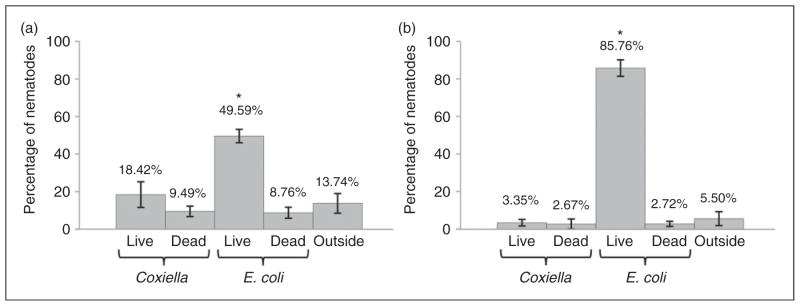
We tested the pathogen avoidance behavior in triplicate populations of 25 wild type nematodes separately transferred to NNGM plates containing four small lawns of different bacterial preparations: (i) live E. coli (CEC), (ii) dead CEC, (iii) live C. burnetii (CCB) and (iv) dead CBB. Both stage 1 and stage 4 larval populations were tested. After 24 h at 25°C, a major percentage of both life-stage nematodes were found on live E. coli, and both life stages appeared to recognize and avoid C. burnetii in a significant manner (P <0.01; Figure 1). Live E. coli OP50 is the standard food for in vitro cultivation of C. elegans, is not recognized as a pathogen and is a preferred nutrient source over dead OP50, live CCB and dead CCB (Figure 1). Furthermore, comparison of L1 (Figure 1a) and L4-stage (Figure 1b) nematodes suggests L4 nematodes have learned to seek live OP50 and avoid potentially noxious (live Coxiella) or nutrient-depleted (autoclaved OP50/Coxiella) food sources.
Figure 1.
Avoidance behavior suggests that C. elegans recognizes C. burnetii as a pathogen. Twenty-five wild type C. elegans stage-1 and stage-4 larval populations were separately transferred (in triplicate) to NNGM plates containing four small lawns of different bacterial preparations: (i) live E. coli (CEC); (ii) dead CEC; (iii) live C. burnetii (CCB); and dead CBB. After 24 h at 25°C, the majority of both: (a) L1-stage and (b) L4-stage nematodes were found on live CEC and both life stages appeared to avoid C. burnetii. All experimental groups were compared by one-way ANOVA (P <0.0001 for L4; P =0.0005 for L1), followed by post-test comparison of all groups by Tukey’s test. Both life stages accumulate on live E. coli in significantly greater numbers than on other food sources (P <0.01).
C. burnetii resists mechanical disintegration
During food ingestion, worms disrupt the meal using a grinder; a specialized organ located in the pharynx that mechanically disintegrates food prior to its entering the intestinal lumen.54 The C. elegans intestine is a tube comprised of 20 epithelial cells called enterocytes, which play roles in nutrient acquisition and immunity.57 Nematodes lack circulating immune cells such as hemocytes of other invertebrates or neutrophils of vertebrates. As such, the nematode intestinal epithelium has evolved to recognize and respond to pathogens in a microbe-specific manner.58
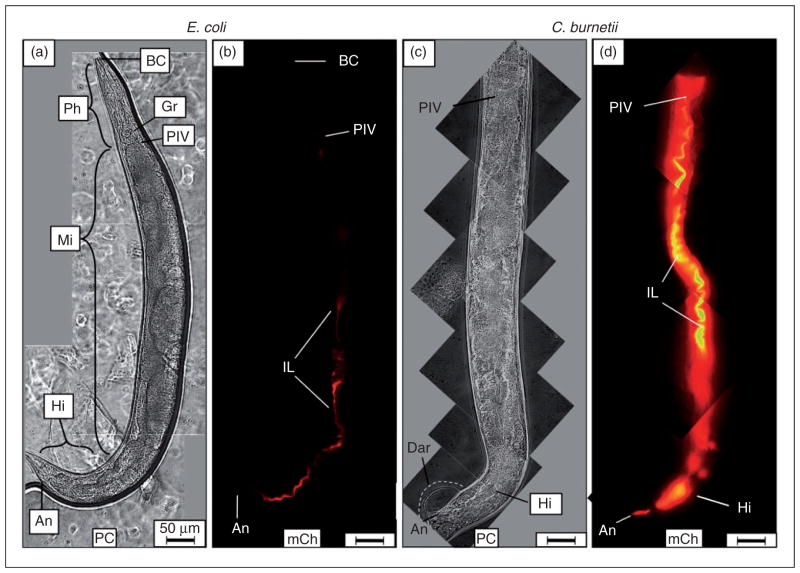
When L4 nematodes were fed live E. coli CEC, intact bacteria were rarely observed in worms at any time point. Instead, the majority of CEC in the intestine appeared to be mechanically and/or enzymatically digested into a homogenous, dim mCherry-containing material (Figure 2a, b). In stark contrast, CCB remained intact after consumption and accumulated in the intestinal lumen of C. elegans with markedly greater mCherry signal intensity (Figure 2c, d). Accumulation initially occurred in two regions of the intestinal lumen, including the anterior portion of the midgut immediately posterior to the pharyngeal-intestinal valve and in the hindgut. Interestingly and coincidentally, these regions of the C. elegans alimentary canal are maintained at a pH of ~4.5,59 an environment well-suited to the acidophilic metabolic activity of C. burnetii.22
Figure 2.
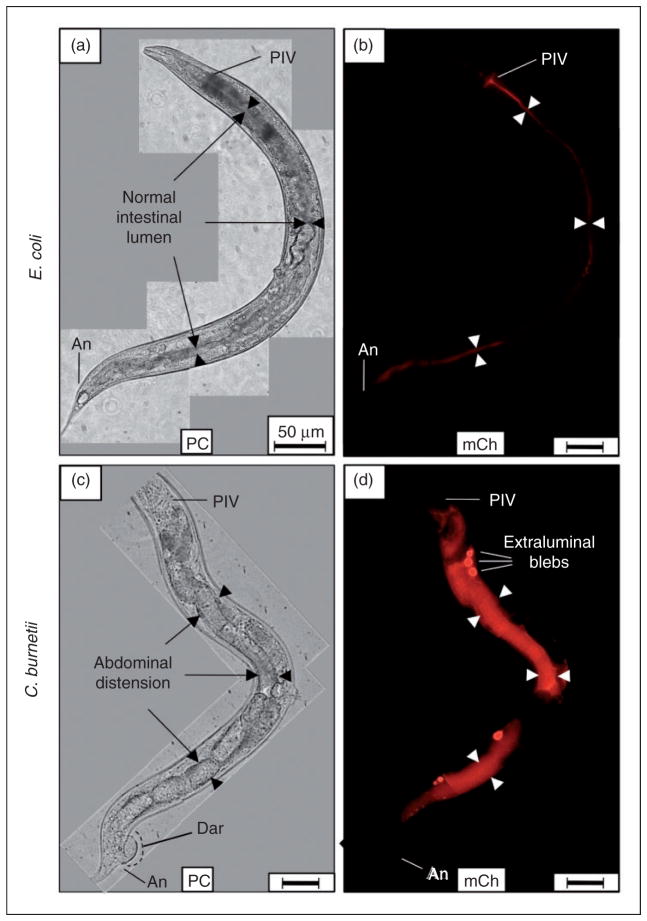
Live C. burnetii accumulates in the intestinal lumen of C. elegans and generates overt pathologies. Wild type C. elegans populations were separately exposed to live, mCherry-expressing- E. coli (CEC) or C. burnetii (CCB) as lawns on the surface of NNGM, and examined by fluorescence microscopy after 4 d of exposure. (a, b) Nematodes fed live CEC consistently showed that C. elegans was capable of digesting live E. coli. (c, d) In contrast, nematodes fed live CCB exhibited an intense mCherry signal that appeared to be restricted to the intestinal lumen. Accumulation of C. burnetii in C. elegans led to a swollen intestinal lumen and Dar that were not observed in worms fed (a) live E. coli or dead C. burnetii (not shown). Images were acquired at the same exposure time and joined and oriented with worm anteriors at the top. Bar =50 μM. BC: buccal cavity; Ph: pharynx; PIV: pharyngeal–intestinal valve; Mi: midgut; Hi: hindgut; An: anus; IL: intestinal lumen; PC: phase contrast; mCh: fluorescent mCherry signal.
The second innate immune defense in C. elegans is the physical barrier created by the exoskeleton and the capacity to mechanically disintegrate ingested microbes processed by the grinding organ. Although we never observed CCB adhering to or entering C. elegans through a compromised exoskeleton, it appeared that Coxiella was capable of subverting the nematode’s second line of defense by remaining intact following passage through the grinder.
Live C. burnetii generates a Dar and intestinal distension in C. elegans
Accumulation of CCB led to several overt pathologies in C. elegans. Perhaps the most striking was a swollen and Dar that grew larger over time (Figures 2c, 3c, and see also Figure 6c–e later). Dar pathology was observed as a swelling of the peri-anal region, where the cuticle was progressively separated from underlying tissue. We were unable to observe mCherry signal within the swollen region, and thus concluded that C. burnetii was absent. The Dar phenotype was not observed in nematodes fed live CEC (Figure 2a) or dead CCB (data not shown), suggesting that metabolic activity of C. burnetii or an autoclave-labile component(s) were responsible for its formation.
Figure 3.
Live C. burnetii accumulates in the intestinal lumen of C. elegans and generates Elbs after 8 d exposure. (a, b) Nematodes fed live E. coli CEC and observed by microscopy after 8 d consistently showed that C. elegans completely digested E. coli without signs of intestinal distension (arrowheads). (c, d) In contrast, nematodes fed live C. burnetii CCB and observed 8 d later exhibited an intense mCherry signal that was mainly restricted to the intestinal lumen. Additionally, Elb pathology was occasionally evident, suggesting colonization of luminal epithelial cells by C. burnetii. Elbs were not evident in (a, b) worms fed live CEC or dead CCB (not shown). Images were acquired at the same exposure time and joined and oriented with worm anteriors at the top. Bar =50 μM. PIV: pharyngeal–intestinal valve; An: anus; PC: phase contrast; mCh: fluorescent mCherry signal.
Live C. burnetii generates extra-luminal bleb pathology in C. elegans
C. burnetii is an obligate intracellular parasite that has been shown to invade and replicate in a wide variety of host cells, including wax moth haemocytes,28 tick tissues,60 alveolar macrophages,61,62 human cardiac valves63 and goat placental trophoblasts.64 Following an 8-d exposure, C. burnetii accumulation in C. elegans occasionally resulted in structures resembling extra-luminal blebs (Elbs; Figure 3d), which were not evident in nematodes that fed on live CEC (Figure 3a, b; Figure 4) or dead CCB (not shown), suggesting that C. burnetii activity or an autoclave-labile metabolite(s) played a role in their formation.
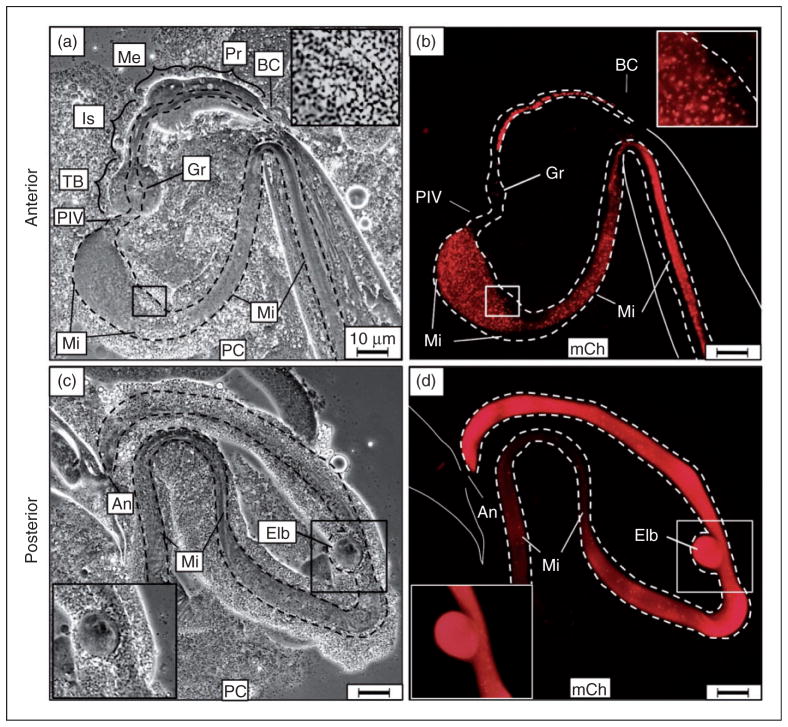
Figure 4.
Examination of anterior and posterior C. elegans extrusions demonstrated intact C. burnetii accumulation in the midgut with extra-intestinal bleb pathology. Following 2-d exposure to live CCB, nematodes were prepared by applying gentle pressure to cover slips with extrusion of intestinal contents through either the mouth (anterior) or anus (posterior). (a, b) Matched panels show large numbers of intact CCB pumped into the pharynx and passed through the grinder into the midgut (insert in b). The extruded intestinal lumen is outlined by dashed lines. (c, d) The posterior extrusion of the nematode indicated intestinal accumulation of CCB in the hindgut with typical Elbs attached and apparently contiguous with the alimentary canal. Images were acquired at same exposure times. Bar =10 μM. BC: buccal cavity; Pr: procorpus; Me: metacorpus; Is: isthmus; TB: terminal bulb; Gr: grinder; PIV: pharyngeal–intestinal valve; An: anus; Mi: midgut; Hi: hindgut; PC: phase contrast; mCh: fluorescent mCherry signal.
Elbs appeared as defined outgrowths or pouches of relatively bright mCherry-labeled material closely associated with the intestinal wall. Elbs varied in size, were observed along the entire length of the intestine, and were distinct from auto-fluorescent lipid material commonly observed in worms as they age. The relatively high-intensity fluorescence of Elbs impaired our ability to visualize individual intact CCB. Elbs may arise from invasion of enterocytes and subsequent formation of parasitophorous vacuole-like microcolonies, as described for Legionella pneumophila infection of C. elegans,65,66 or accumulation in coelomocyte vesicles.67
To further characterize Elbs, anterior and posterior portions of infected alimentary canals were examined. Application of gentle pressure to worms forced the intestine and attached tissues to extrude outside the body through either the mouth (anterior) or anus (posterior). Analysis of the anterior-extruded contents of infected worms demonstrated the presence of intact CCB in the procorpus, metacorpus and isthmus of the pharynx, as well as following passage through the terminal bulb grinder and into the midgut (Figure 4a, b).
Transmission electron microscopy demonstrates accumulation of intact C. burnetii in C. elegans
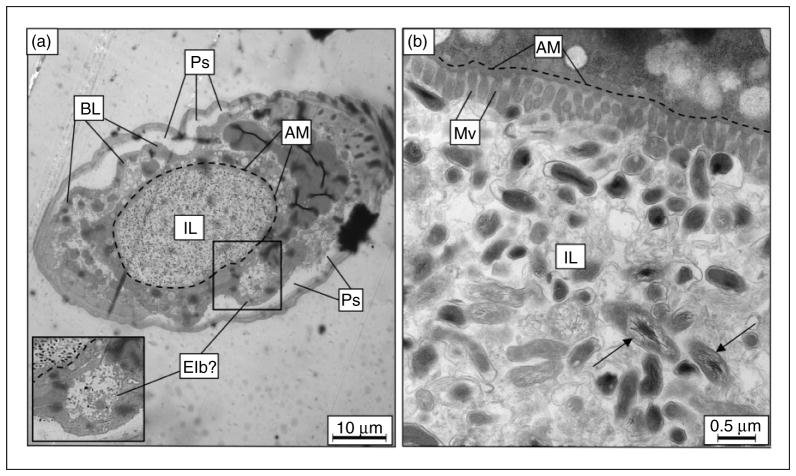
To more closely examine a C. burnetii infection of C. elegans, L4 nematodes were fed live CCB for 6 d, transverse cross-sectioned, and examined by TEM. TEM analysis at low magnification (1500×) demonstrated that the intestinal lumen was packed with intact C. burnetii and other amorphous material (Figure 5a). Although we did not examine E. coli-exposed worms, a previous study comparing E. coli OP50, Pseudomonas aeruginosa and Staphylococcus aureus infections of C. elegans used TEM to show that S. aureus generates intestinal distension and enterocyte effacement,68 similar to what was observed in C. burnetii-infected C. elegans (Figure 5b). Although further analysis is needed to more fully characterize Elb outgrowths of the intestinal lumen, we observed structures by TEM that were consistent with these blebs in several specimens (Figure 5a).
Figure 5.
TEM of C. elegans fed live C. burnetii. After 6 d on a live C. burnetii diet, cross-sections of infected nematodes (a) at low magnification (1,500×) showed a distended intestinal lumen packed with bacteria. Apical membranes of two enterocytes delineate the intestinal lumen boundary, here outlined with a dashed line, showing intestinal distension. A structure resembling an Elb is boxed and magnified as inset. (b) At higher magnification (20,000×), numerous intact C. burnetii were observed in intimate contact with enterocytes, and the microvillar boarder appeared to be shortened and effaced. C. burnetii with distended nucleoids (arrows) were an indicator of C. burnetii vegetative cells, and were intact within the intestinal lumen. IL: intestinal lumen; Ps: pseudocoelom; BL: basal lamina; AM: apical membrane; Mv: microvilli.
At higher magnification (20,000×), TEM revealed an intimate relationship between intact C. burnetii and the microvillar border of the apical membrane (Figure 5b). As mentioned, S. aureus infection results in effacement of C. elegans enterocytes,68 indicated by a loss of integrity in the microvillar border and shortened length of individual microvilli; similar to what we observed in C. burnetii-infected worms. An amorphous substance was also seen in the lumen with the intact bacteria and may represent nematode glycocalyx secretions. This intimate host–pathogen contact is possibly involved in pathologies exhibited by CCB-infected C. elegans. In fact, the enterocytes in infected worms appeared unhealthy, where a decreased number of secretory vesicles along with nuclear blebbing/condensed chromatin suggested that cells may be undergoing apoptosis.69
C. elegans used in this experiment were fed 7–10-d-old ACCM-2-grown CCB that contained a mixed population of SCVs and LCVs. Chromosomal (nucleoid) dispersion has previously been used as an indicator of metabolic activity in LCVs following growth in axenic culture.70,71 Although further analyses are required to characterize the metabolic activity of C. burnetii inside C. elegans, the presence of numerous bacteria with dispersed nucleoids strongly suggests that the more fragile and metabolically active LCVs were present.
C. burnetii infection decreases the lifespan of C. elegans
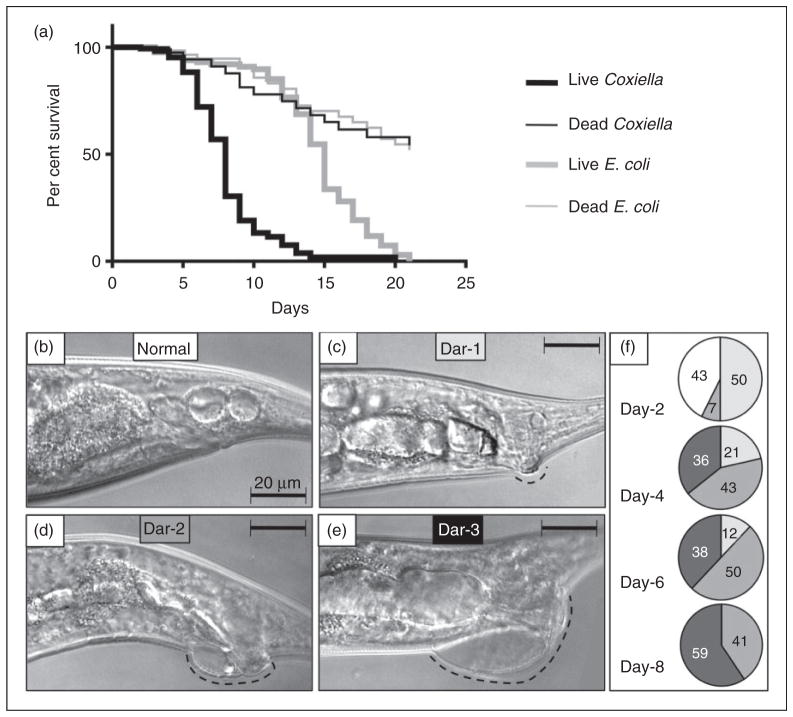
Analysis of the C. elegans lifespan is one of the most common quantitative methods used to characterize the effects of genetic and biological factors associated with the process of aging,72 and to assay ‘slow’ killing pathologies exhibited by C. elegans following various microbial challenges. In contrast to the relatively fast death (1–2 d) exhibited by young larvae upon exposure to live Coxiella (data not shown), L4-stage nematodes lived much longer and exhibited a ‘slow’ killing pathology, indicated by a shortened lifespan.73 Thus, we assessed survival of wild type N2 C. elegans exposed to CCB or E. coli OP50 that were either live or killed by autoclaving. Each of the four test treatments was provided as a lawn on the surface of NNGM plates, and 30 L4 nematodes were initially exposed to each of the four treatments in quintuplicate. Live worms exposed to each of the four treatments were enumerated daily for 20 d and transferred each day to fresh NNGM plates containing the same bacteria to generate survival curves (Figure 6a).
While exposure to live E. coli OP50 did not induce overt pathologies in C. elegans, a reduced lifespan (15-d median) compared with worms exposed to dead E. coli (>21-d median) suggests that the standard bacterial food source elicits a slow killing pathology, as previously reported.42 Interestingly, the median life spans of nematodes were not significantly different (P =0.9014) when fed dead E. coli or dead C. burnetii, suggesting that either bacterium can serve as a food source. In contrast, a significant difference (P <0.0001) was observed between worms fed live E. coli or live CCB, where mean survival times were 15 d and 8 d, respectively (Figure 6a).
Figure 6.
Survival assays show C. burnetii infection results in a significantly reduced lifespan of C. elegans concurrent with progressively increased Dar pathology. (a) Kaplan–Meier survival plots of N2 nematodes fed live/dead E. coli OP50 or live/dead CCB and observed every 24 h. Worms were classified as live if motile or responsive to touch. Dead worms were non-motile and non-responsive to touch. Worms that left NNGM plates were censored from the assay. Median life spans of nematodes were not significantly different (P =0.9014 by pair-wise log-rank test comparison) when fed dead E. coli or dead C. burnetii. However, a significant difference (P <0.0001 by pair-wise log-rank test comparison) was observed between worms fed live E. coli or live C. burnetii, with mean survival times of 15 d and 8 d, respectively. (b) Initially, Dar pathology (dashed line) was not evident, but, over time, progressively greater Dar pathologies (c, Dar-1 (weak); d, Dar-2 (moderate); e, Dar-3 (severe), were observed. (f) Percentage of worms with Dar pathologies as a function of days post-infection. All nematodes exhibited ≥Dar-2 by 4 d. By 8 d 59% of nematodes exhibited Dar-3. Images were acquired by differential interference microscopy. Bar =20 μM.
To better understand Dar pathology, we assessed the size and prevalence of the peri-anal swellings induced by C. burnetii over time (Figure 6b–f). Worms initially showed a normal peri-anal region (Figure 6b), but by 2 d and beyond, lesions increased in frequency and prominence (Figure 6c–e). The percentage of C. elegans displaying each Dar type (Dar-1, weak; Dar-2, moderate; Dar-3, severe) was assessed over time (Figure 6f), and although no worms had Dar on d 0, by d 4 all worms had some degree of peri-anal swelling. By d 8, 59% of CBB-infected C. elegans exhibited severe swelling (Dar-3).
Lifespan of C. burnetii-infected C. elegans TLR mutant tol-1(−) is not significantly different from wild type C. elegans
Up to this point, the results of the study clearly demonstrated a number of pathologies (e.g. Dar, intestinal distension, Elbs, decreased lifespan) in C. elegans following ingestion of live CCB that were not apparent after exposure to live E. coli or dead CCB. These pathologies suggested that the nematode’s innate immune system might be activated in a fashion similar to the responses elicited by challenge with Microbacterium nematophilum or S. aureus. Thus, further exploration of C. elegans as a model for studying the molecular biology of C. burnetii pathogenesis and host immune response was warranted.
Host cells utilize pattern recognition receptors (PRRs) to detect microbial PAMPs and activate a particular subset of immunomodulatory mediators to deal with a specific pathogen.74–76 The Drosophila melanogaster Toll-1 receptor was one of the first studied PRRs; initially it was identified as being required for proper embryonic development,77 and later shown to play a significant role in the innate immune response of the fruit fly.78 Subsequently, eight additional Toll-1 homologues, collectively referred to as TLRs were identified in D. melanogaster and 11 have been described, to date, in mammals.74,78
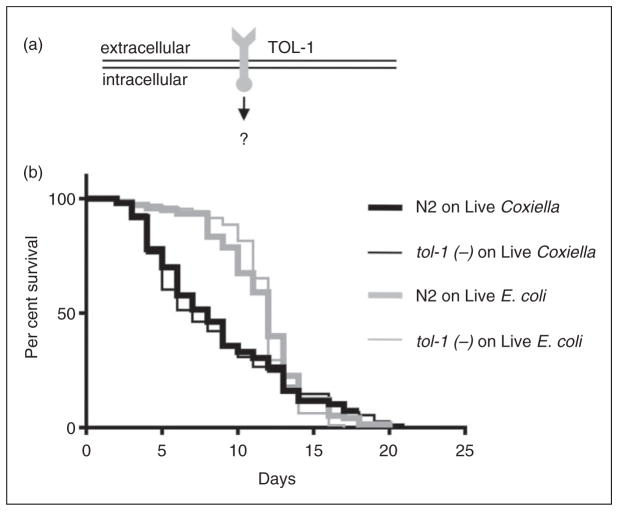
Several PRRs have been identified in C. elegans,79,80 including a single Toll-1 ortholog, termed TOL-1 (Figure 7a). Although the signaling cascade(s) associated with this PRR is unclear, we asked whether C. elegans uses TOL-1 to detect and respond to C. burnetii by comparing pathological phenotypes and longevity of wild type and tol-1 mutant C. elegans fed live E. coli OP50 or CCB. As observed before, when wild type C. elegans were exposed to live CCB, their life spans were significantly (P <0.0009 by pair-wise log-rank test) decreased relative to live E. coli (Figure 7b). However, we observed no significant differences in survival between wild type and tol-1 mutants fed live CCB (P =0.6787 by pair-wise log-rank test; Figure 7b), nor were there any observable differences in frequency or severity of Dar pathology or intestinal distension (data not shown). These results strongly suggest that C. elegans TOL-1 does not directly participate in the host response to C. burnetii.
Figure 7.
Deletion of the TLR, TOL-1, does not affect the lifespan of C. elegans challenged with C. burnetii. (a) TLRs are used by vertebrates for differential detection of various microbes, including C. burnetii. Although the associated signaling cascade(s) is unclear, C. elegans has a single TLR (TOL-1) orthologue. (b) Kaplan–Meier survival plot of wild type or tol-1(−) C. elegans exposed to live E. coli OP50 or CCB. Survival analyses demonstrate that the survival curves of tol-1(−) nematodes exposed to live E. coli or CCB were significantly different (P =0.0015 by pair-wise log-rank test) with median survival times decreased from 12 d (when fed live E. coli) to 7 d (when fed live CCB). However, there was no significant difference (P =0.6787 by pair-wise log-rank test) in the life spans of tol-1(−) compared with wild type (WT) N2 nematodes exposed to live CCB or live E. coli OP50, suggesting that TOL-1 is not used to detect C. burnetii.
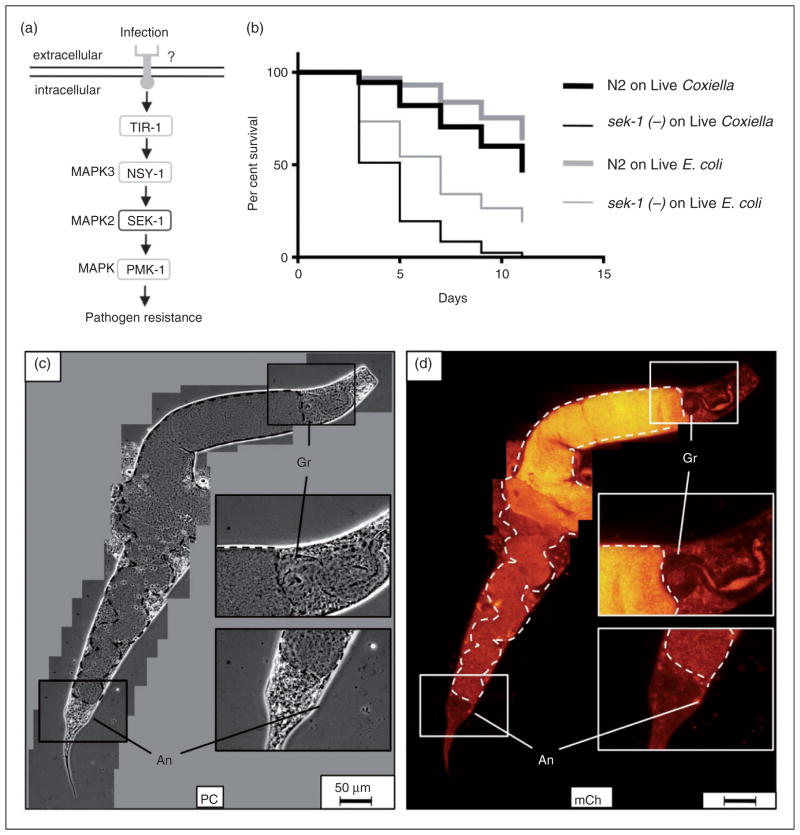
C. elegans sek-1(−) immune signaling mutants exhibit a significantly reduced lifespan and pronounced intestinal accumulation of C. burnetii
A genetic screen for C. elegans mutants that were hyper-susceptible to P. aeruginosa killing81 identified SEK-1 and NSY-1, two MAPKs (Figure 8a adapted from37) involved in cell-fate decision and required for nematode resistance to P. aeruginosa.37 Although an associated PRR(s) has yet to be identified, SEK-1 and NSY-1 are components of a MAPK signaling pathway immediately upstream from PMK-1, one of three known p38-family MAPKs in C. elegans.37 Previous studies have also shown that MAPKs are utilized by vertebrates to signal the presence of C. burnetii.61,82–85 Furthermore, MAPKs of Saccharomyces cerevisiae yeast are modulated by C. burnetii’s Icm/Dot type IVB secretion system effector proteins.86
Figure 8.
Lifespan of C. elegans sek-1(−) is significantly decreased and accompanied by markedly overt intestinal distension and live C. burnetii accumulation. (a) SEK-1 is a MAPK that participates in P. aeruginosa infection of nematodes through an unknown receptor(s) (adapted from Ewbank37). (b) Kaplan–Meier survival plot of wild type or sek-1(−) C. elegans exposed to live E. coli OP50 or CCB. Survival assays comparing sek-1 mutants exposed to live E. coli or CCB showed a significant (P <0.0001) lifespan reduction with median survival times of 7 d on live E. coli and 5 d on live C. burnetii. (c, d) Fluorescence microscopy suggests that premature death of sek-1(−) mutants is associated with pronounced intestinal distension (intestinal lumen outlined with dashed line). Dar pathologies were observed but rare (not shown). SEK-1 nematode imaged after 9 d of CCB exposure. Bar =50 μM. Gr: grinder; An: anus; PC: phase contrast; mCh: fluorescent mCherry signal.
For these reasons, we analyzed C. elegans sek-1 mutants to characterize the role of this MAPK signaling pathway in response to C. burnetii exposure. To accomplish this, we compared longevities of wild type and sek-1(−) C. elegans exposed to live CCB and E. coli OP50 (Figure 8b). Sek-1(−) nematodes exposed to live E. coli or CCB exhibited significant decreases in survival as compared to wild type nematodes subjected to the same treatments (P <0.0001 by pair-wise log-rank test for each treatment). Furthermore, exposure of sek-1(−) mutants to live CCB resulted in a significant (P <0.0001) lifespan reduction (5 d median survival time) relative to exposure to live E. coli (7 d median survival time) (Figure 8b).
CCB infection of sek-1 mutants was also characterized by fluorescence microscopy, and the most overt pathology was remarkable intestinal distension, the greatest degree seen in any nematode strain in this study (Figure 8c, d). C. elegans became packed with CCB and after 9 d, the intestinal lumen appeared to span the entire width of the nematode. Moreover, the hindgut region of the intestinal canal often appeared coiled owing to swelling from accumulation of CCB (Figure 8c). While CCB-infected sek-1 mutants exhibited Dar, this pathology was relatively decreased in frequency and severity compared with infected wild type or tol-1(−) nematodes. For example, by 8 d all infected wild type nematodes exhibited ≥Dar-2 swelling (see Figure 6e), whereas even after 9 d a Dar phenotype was rarely observed in sek-1(−) mutants (Figure 8c). These results suggest that the C. elegans immune signaling response to C. burnetii involves MAPK SEK-1.
C. elegans daf-2(ts) mutants are colonized by Coxiella but exhibit diminished pathologies relative to wild type nematodes
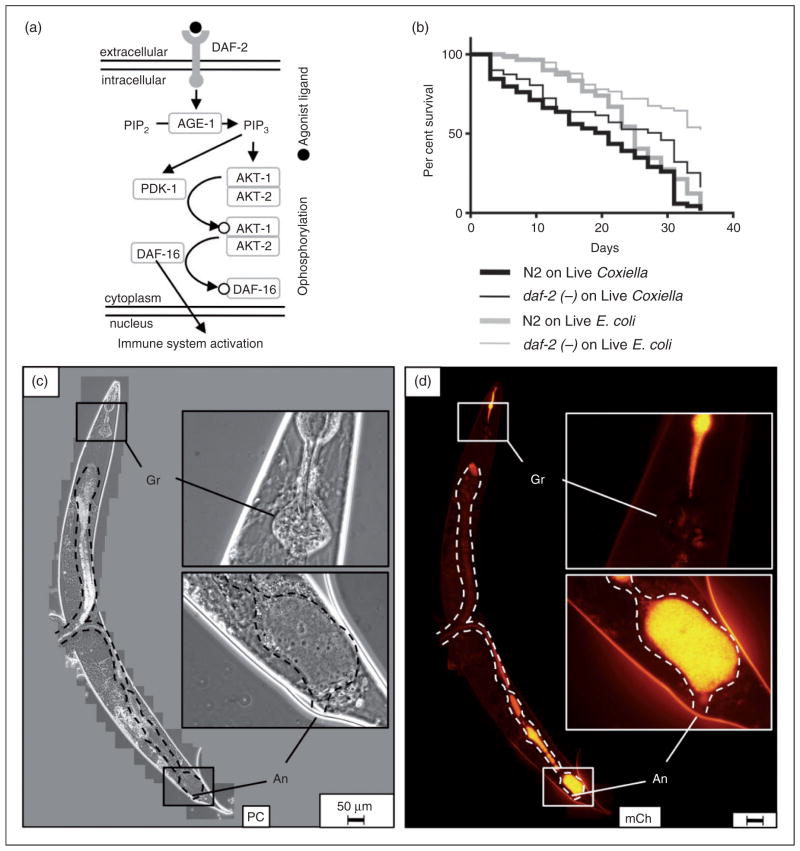
The C. elegans lysozyme gene lys-8 is up-regulated upon bacterial infection, and both DAF-2/DAF-16 (insulin-receptor-like) and DBL-1/TGF-β signaling pathways have been shown to participate in this process.37,87 The DAF-2/DAF-16 pathway is activated when an agonist binds to the insulin-like growth factor DAF-2 membrane receptor tyrosine kinase and initiates a phosphorylation cascade, ultimately resulting in cytoplasmic retention of DAF-16-P (Figure 9a). However, when DAF-2 is bound by an antagonist or the corresponding gene is mutated, DAF-16 is not phosphorylated and instead translocates to the nucleus where it up-regulates a number of antimicrobial and stress-response genes. Thus, daf-2 mutants are more resistant to infection by a variety of pathogenic bacteria owing to DAF-16-induced immune system activation and production of antimicrobials, such as lysozyme.37,88
Figure 9.
Caenorhabditis elegans daf-2 mutants survive significantly longer and with less pathological consequences than wild type nematodes during a C. burnetii infection. (a) Caenorhabditis elegans DAF-2/DAF16 pathway is activated when an agonist binds to DAF-2 receptor resulting in cytoplasmic retention of DAF-16-P, whereas in DAF-2 mutants, nuclear DAF-16 translocation results in activation of immune-related genes (adapted from Ewbank37). (b) Kaplan–Meier survival plot of wild type or daf-2(ts) C. elegans exposed to live E. coli OP50 or CCB. Survival assays demonstrated a significant difference between median survival of wild type C. elegans and daf-2 mutants exposed to CCB (P =0.002 by pair-wise log-rank comparison) and suggest that nematodes are capable of controlling a Coxiella infection. The assay was performed at 20°C. (c, d) Fluorescence microscopy of daf-2 mutants revealed minimal intestinal distension (intestinal lumen-dashed outline), decreased bacterial load per worm and hindgut colonization by inspection. daf-2(ts) nematode imaged after 33 d of exposure to live CCB. Bar =50 μM. Gr: grinder; An: anus; PC: phase contrast; mCh: fluorescent mCherry signal.
Thus, we were curious whether C. elegans daf-2 mutants were also capable of resisting a C. burnetii infection. Survival of daf-2(ts) and wild type nematodes was analyzed in parallel upon exposure to either live CCB or E. coli OP50 (Figure 9b). A significant (P =0.0023 by pair-wise log-rank test) difference between survival curves upon exposure to CCB was observed, with median survival of wild type C. elegans at 21 d and DAF-2 mutant at 29 d, suggesting that a daf-2 mutation aids in controlling Coxiella infection. Analysis of CCB-infected daf-2 mutants by fluorescence microscopy showed a marked decrease in intestinal distension and bacterial load per worm by visual inspection (Figure 9c, d). Intact CCB appeared to accumulate in the hindgut of daf-2 mutants, and although Dar pathologies were occasionally observed, they were restricted to ≤Dar-1 (not shown).
Discussion
Compared with vertebrate host–pathogen model systems, C. elegans provides a number of advantages for studying the innate immune response to C. burnetii: (i) C. elegans is relatively inexpensive to maintain, house, and manipulate; (ii) large populations of hosts can be generated in a relatively short amount of time, as a single adult C. elegans can lay >200 eggs which hatch and develop into egg-laying adults in just a few days; (iii) C. elegans bodies are transparent, which allows easy microscopic visualization of host–pathogen interactions over time in live animals; and (iv) C. elegans are genetically tractable and mutants in nearly every genetic locus are available for study.
Our results demonstrate that C. elegans is capable of recognizing and avoiding C. burnetii, which is the nematode’s first line of innate immunity. This avoidance behavior has been attributed, in part, to the worm’s ability to sense its environment via NPR-1, a homologue of the mammalian neuropeptide Y receptor,89 as well as a ubiquitin ligase.90 It would be interesting to study these mutants to understand the molecular basis of the Coxiella avoidance behavior.
Dar is a C. elegans pathology first described as a result of infection by M. nematophilum.91 Several laboratories independently observed Dar in C. elegans cultures and initially assumed it was the result of a naturally occurring mutation(s). It was later determined that Dar was actually the result of an infection by M. nematophilum, a new species of a Gram-positive coryneform bacterium fortuitously isolated in contaminated C. elegans cultures.91 M. nematophilum adheres to rectal epithelium and post-anal cuticle, where it induces local swelling and constipation that leads to a non-lethal, slowed-growth phenotype.91
Although diverse bacteria are pathogens of C. elegans,92 the pathologies and innate immune response induced by M. nematophilum are somewhat unusual and are reminiscent of what we observed during a C. burnetii infection (Figures 2, 3 and 6). C. elegans nucleotide sugar transporter mutants (srf-3) were previously shown to be resistant to a M. nematophilum infection, as well as to Yersinia adherence and biofilm formation.93,94 A srf-3 mutation resulted in a 65% reduction in C. elegans O-linked glycoconjugates, strongly suggesting that secreted nematode glycoconjugates play a role in adherence of M. nematophilum and Yersinia to the C. elegans intestinal lumen.93,94 Thus, future studies with C. elegans srf-3 mutants may indicate a similar immune mechanism during a C. burnetii infection.
Previously, chemical and transposon libraries of C. elegans were screened for bacterial un-swollen (bus) mutants that exhibited little or no Dar following a M. nematophilum infection to identify genes involved in bacterial adhesion/colonization or the swelling response,95 including c-type lectins,96 galactosyltransferase,97,98 mucins99 and O-glycoconjugates.100 One of the isolated C. elegans mutants (bus-4) was resistant to infection by M. nematophilum and Yersinia owing to overproduction of a nematode mucin equivalent.99 Interestingly, surfactant protein D is a C-type lectin and innate immune effector present in many vertebrate mucosal surfaces such as the lung, and has recently been shown to bind C. burnetii and participate in subversion of phagocytosis in mouse alveolar macrophages.101 Future C. burnetii infection studies involving bus-4(−) mutant strains will test whether a similar mechanism may mediate resistance to C. burnetii infection.
In this study, we analyzed the ability of C. burnetii RSA 439 (phase II) to infect C. elegans and demonstrated a lack of participation by TOL-1, a TLR, in the nematode’s response to the bacterium (Figure 7). A number of studies have investigated the role of TLRs in the detection and immune response to C. burnetii, and results strongly suggest that TLR1,30,32 TLR2102–105 and TLR431,106–108 participate in pathogen recognition and signaling, and that bacterial LPS plays a pivotal role in recognition shielding. When wild type, virulent C. burnetii (phase I) is passaged in vitro, naturally occurring mutation(s) result in loss of genes required for synthesis of full-length LPS with a concurrent loss of virulence. These attenuated LPS mutant strains are referred to as phase II.109,110 It is thought that phase I LPS shields C. burnetii from detection by host Abs,111 complement112 and TLRs,113 and thus enables C. burnetii infection to progress. As we used C. burnetii phase II in this study, it would be interesting to understand how C. elegans responds to full-length C. burnetii LPS and virulent phase I C. burnetii. However, our results demonstrate activation of the C. elegans immune system by TLR/LPS-independent mechanism(s), suggesting that alternate evolutionarily conserved pathways such as TFEB/HLH-3039 may play a role in vertebrate innate immune recognition of C. burnetii.
CCB-infected sek-1(−) mutants exhibited Dar (Figure 8) with a markedly decreased frequency and severity relative to CCB-infected wild type nematodes (Figure 6). It is logical to hypothesize that this may be owing to SEK-1-mediated signaling being essential for the worm’s pathogen invasion response manifested by swelling/Dar or overlap in signaling outcomes with other pathways such as ERK.37,114 We also showed that DAF-2 mutants were resistant to infection by CCB, likely owing to overproduction of antimicrobials such as lysozyme.37,88 Future studies characterizing invertebrate signaling and effector antimicrobials involved might eventually lead to novel approaches for developing therapeutics for infected humans and domesticated ruminants.
In this study, we describe several overt manifestations in C. elegans responding to a C. burnetii exposure, including pathogen avoidance and pathological results of infection (Dar, a significantly reduced longevity, Elbs and intestinal distension), which indicate activation of the nematode’s inducible immune system. These features make this model particularly attractive for investigating Coxiella’s molecular pathogenesis. In conclusion, our results demonstrate the utility of C. elegans as a user-friendly model for studying Coxiella virulence and host innate immunity mechanisms. Currently, we are attempting to characterize C. burnetii molecule(s) that play a role in inducing the immune system of C. elegans (C. burnetii Tn-seq assay),115 as well as to better understand the specific induced innate immune response mechanisms of the nematode (transcriptomic and proteomic analyses) and identify potential homologues in humans.
Acknowledgments
We thank Margie Kinnersley, Bill Granath, Jim Driver, Indu Warrier, Carson Whitehill, Mary Ellenbecker, Hannah Fay, Xiaobo Wang, Julia Battisti, (University of Montana) and Ann-Karen Brassinga (University of Manitoba) for technical assistance and useful discussions related to this project.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH-NIAID grants R15 AI103511 (to JMB) and R21 AI123293 (to MFM).
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Maurin M, Raoult D. Q fever. Clin Microbiol Rev. 1999;12:518–553. doi: 10.1128/cmr.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duron O, Sidi-Boumedine K, Rousset E, et al. The importance of ticks in q fever transmission: what has (and has not) been demonstrated? Trends Parasitol. 2015;31:536–552. doi: 10.1016/j.pt.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Georgiev M, Afonso A, Neubauer H, et al. Q fever in humans and farm animals in four European countries, 1982 to 2010. Euro Surveill. 2013;18:1–13. [PubMed] [Google Scholar]
- 4.Van den Brom R, van Engelen E, Roest HI, et al. Coxiella burnetii infections in sheep or goats: an opinionated review. Vet Microbiol. 2015;181:119–129. doi: 10.1016/j.vetmic.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Meredith AL, Cleaveland SC, Denwood MJ, et al. Coxiella burnetii (Q-fever) seroprevalence in prey and predators in the United Kingdom: evaluation of infection in wild rodents, foxes and domestic cats using a modified ELISA. Transbound Emerg Dis. 2015;62:639–649. doi: 10.1111/tbed.12211. [DOI] [PubMed] [Google Scholar]
- 6.Joulie A, Laroucau K, Bailly X, et al. Circulation of Coxiella burnetii in a naturally Infected Flock of Dairy Sheep: Shedding Dynamics, Environmental Contamination, and Genotype Diversity. Appl Environ Microbiol. 2015;81:7253–7260. doi: 10.1128/AEM.02180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro AJ, Norris JM, Heller J, et al. Seroprevalence of Coxiella burnetii in Australian dogs. Zoonoses Public Health. 2016;63:458–466. doi: 10.1111/zph.12250. [DOI] [PubMed] [Google Scholar]
- 8.Agerholm JS. Coxiella burnetii associated reproductive disorders in domestic animals—a critical review. Acta Vet Scand. 2013;55:13. doi: 10.1186/1751-0147-55-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson AD, Szymanski TJ, Emery MP, et al. Epizootiological investigation of a Q fever outbreak and implications for future control strategies. J Am Vet Med Assoc. 2015;247:1379–1386. doi: 10.2460/javma.247.12.1379. [DOI] [PubMed] [Google Scholar]
- 10.Anderson A, Bijlmer H, Fournier PE, et al. Diagnosis and management of Q fever—United States, 2013: recommendations from CDC and the Q Fever Working Group. MMWR Recomm Rep. 2013;62:1–30. [PubMed] [Google Scholar]
- 11.Angelakis E, Raoult D. Q Fever. Vet Microbiol. 2010;140:297–309. doi: 10.1016/j.vetmic.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Mares-Guia MA, Rozental T, Guterres A, et al. Molecular identification of Q fever in patients with a suspected diagnosis of dengue in Brazil in 2013–2014. Am J Trop Med Hyg. 2016;94:1090–1094. doi: 10.4269/ajtmh.15-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonso E, Lopez-Etxaniz I, Hurtado A, et al. Q fever outbreak among workers at a waste-sorting plant. PLOS ONE. 2015;10:e0138817. doi: 10.1371/journal.pone.0138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snedeker KG, Sikora C. Q fever in Alberta, Canada: 1998–2011. Zoonoses Public Health. 2014;61:124–130. doi: 10.1111/zph.12053. [DOI] [PubMed] [Google Scholar]
- 15.Szymanska-Czerwinska M, Galinska EM, Niemczuk K, Knap JP. Prevalence of Coxiella burnetii infection in humans occupationally exposed to animals in Poland. Vector Borne Zoonotic Dis. 2015;15:261–267. doi: 10.1089/vbz.2014.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noda AA, Rodriguez I, Miranda J, et al. First molecular evidence of Coxiella burnetii infecting ticks in Cuba. Ticks Tick Borne Dis. 2016;7:68–70. doi: 10.1016/j.ttbdis.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Bontje DM, Backer JA, Hogerwerf L, et al. Analysis of Q fever in Dutch dairy goat herds and assessment of control measures by means of a transmission model. Prev Vet Med. 2016;123:71–s89. doi: 10.1016/j.prevetmed.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Biggs HM, Turabelidze G, Pratt D, et al. Coxiella burnetii infection in a community operating a large-scale cow and goat dairy, Missouri, 2013. Am J Trop Med Hyg. 2016;94:525–531. doi: 10.4269/ajtmh.15-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Hoek W, Morroy G, Renders NH, et al. Epidemic Q fever in humans in the Netherlands. Adv Exp Med Biol. 2012;984:329–364. doi: 10.1007/978-94-007-4315-1_17. [DOI] [PubMed] [Google Scholar]
- 20.Schneeberger PM, Wintenberger C, van der Hoek W, Stahl JP. Q fever in the Netherlands—2007–2010: what we learned from the largest outbreak ever. Med Mal Infect. 2014;44:339–353. doi: 10.1016/j.medmal.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Hogerwerf L, van den Brom R, Roest HI, et al. Reduction of Coxiella burnetii prevalence by vaccination of goats and sheep, The Netherlands. Emerg Infect Dis. 2011;17:379–386. doi: 10.3201/eid1703.101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voth DE, Heinzen RA. Lounging in a lysosome: the intra-cellular lifestyle of Coxiella burnetii. Cell Microbiol. 2007;9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 23.van Loenhout JA, Hautvast JL, Vercoulen JH, et al. Q-fever patients suffer from impaired health status long after the acute phase of the illness: results from a 24-month cohort study. J Infect. 2015;70:237–246. doi: 10.1016/j.jinf.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Harris RJ, Storm PA, Lloyd A, et al. Long-term persistence of Coxiella burnetii in the host after primary Q fever. Epidemiol Infect. 2000;124:543–549. doi: 10.1017/s0950268899003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Million M, Thuny F, Richet H, Raoult D. Long-term outcome of Q fever endocarditis: a 26-year personal survey. Lancet Infect Dis. 2010;10:527–535. doi: 10.1016/S1473-3099(10)70135-3. [DOI] [PubMed] [Google Scholar]
- 26.van der Hoek W, Versteeg B, Meekelenkamp JC, et al. Follow-up of 686 patients with acute Q fever and detection of chronic infection. Clin Infect Dis. 2011;52:1431–1436. doi: 10.1093/cid/cir234. [DOI] [PubMed] [Google Scholar]
- 27.Bewley KR. Animal models of Q fever (Coxiella burnetii) Comp Med. 2013;63:469–476. [PMC free article] [PubMed] [Google Scholar]
- 28.Norville IH, Hartley MG, Martinez E, et al. Galleria mellonella as an alternative model of Coxiella burnetii infection. Microbiology. 2014;160:1175–1181. doi: 10.1099/mic.0.077230-0. [DOI] [PubMed] [Google Scholar]
- 29.Capo C, Mege JL. Role of innate and adaptive immunity in the control of Q fever. Adv Exp Med Biol. 2012;984:273–286. doi: 10.1007/978-94-007-4315-1_14. [DOI] [PubMed] [Google Scholar]
- 30.Ammerdorffer A, Schoffelen T, Gresnigt MS, et al. Recognition of Coxiella burnetii by toll-like receptors and nucleotide-binding oligomerization domain-like receptors. J Infect Dis. 2015;211:978–987. doi: 10.1093/infdis/jiu526. [DOI] [PubMed] [Google Scholar]
- 31.Ramstead AG, Robison A, Blackwell A, et al. Roles of TLR2, TLR4, and MyD88 during pulmonary Coxiella burnetii infection. Infect Immun. 2016;84:940–949. doi: 10.1128/IAI.00898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoffelen T, Ammerdorffer A, Hagenaars JC, et al. Genetic variation in pattern recognition receptors and adaptor proteins associated with development of chronic Q fever. J Infect Dis. 2015;212:818–829. doi: 10.1093/infdis/jiv113. [DOI] [PubMed] [Google Scholar]
- 33.Schoffelen T, Wegdam-Blans MC, Ammerdorffer A, et al. Specific in vitro interferon-gamma and IL-2 production as bio-markers during treatment of chronic Q fever. Front Microbiol. 2015;6:93. doi: 10.3389/fmicb.2015.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neher DA. Ecology of plant and free-living nematodes in natural and agricultural soil. Annu Rev Phytopathol. 2010;48:371–394. doi: 10.1146/annurev-phyto-073009-114439. [DOI] [PubMed] [Google Scholar]
- 35.Engelmann I, Pujol N. Innate immunity in C. elegans. Adv Exp Med Biol. 2010;708:105–121. doi: 10.1007/978-1-4419-8059-5_6. [DOI] [PubMed] [Google Scholar]
- 36.Li Q, Liberles SD. Aversion and attraction through olfaction. Curr Biol. 2015;25:R120–R129. doi: 10.1016/j.cub.2014.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewbank JJ. Signaling in the immune response. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.83.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 38.Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr Opin Immunol. 2012;24:3–9. doi: 10.1016/j.coi.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Visvikis O, Ihuegbu N, Labed SA, et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sifri CD, Begun J, Ausubel FM. The worm has turned–microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Corsi AK. A biochemist’s guide to Caenorhabditis elegans. Anal Biochem. 2006;359:1–17. doi: 10.1016/j.ab.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darby C. Interactions with microbial pathogens. WormBook. 2005:1–15. doi: 10.1895/wormbook.1.21.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 43.Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol Biol. 2008;415:403–427. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
- 44.Hinnebusch BJ, Rudolph AE, Cherepanov P, et al. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 45.Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- 46.Omsland A, Beare PA, Hill J, et al. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol. 2011;77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C, Banga S, Mertens K, et al. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A. 2010;107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber MM, Chen C, Rowin K, et al. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol. 2013;195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beare PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol. 2012;78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beare PA. Genetic manipulation of Coxiella burnetii. Adv Exp Med Biol. 2012;984:249–271. doi: 10.1007/978-94-007-4315-1_13. [DOI] [PubMed] [Google Scholar]
- 51.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 52.Shaham S. WormBook. 2006. The C. elegans Research Community. [Google Scholar]
- 53.Hall DH, Hartwieg E, Nguyen KC. Modern electron microscopy methods for C. elegans. Methods Cell Biol. 2012;107:93–149. doi: 10.1016/B978-0-12-394620-1.00004-7. [DOI] [PubMed] [Google Scholar]
- 54.Avery L, You YJ. C. elegans feeding. WormBook. 2012:1–23. doi: 10.1895/wormbook.1.150.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 55.Bargmann CI. Chemosensation in C. elegans. WormBook. 2006:1–29. doi: 10.1895/wormbook.1.123.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 56.Schulenburg H, Ewbank JJ. The genetics of pathogen avoidance in Caenorhabditis elegans. Mol Microbiol. 2007;66:563–570. doi: 10.1111/j.1365-2958.2007.05946.x. [DOI] [PubMed] [Google Scholar]
- 57.McGhee JD. The C. elegans intestine. WormBook. 2007:1–36. doi: 10.1895/wormbook.1.133.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 58.Schulenburg H, Hoeppner MP, Weiner J, 3rd, Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 2008;213:237–250. doi: 10.1016/j.imbio.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Chauhan VM, Orsi G, Brown A, et al. Mapping the pharyngeal and intestinal pH of Caenorhabditis elegans and real-time luminal pH oscillations using extended dynamic range pH-sensitive nanosensors. ACS Nano. 2013;7:5577–5587. doi: 10.1021/nn401856u. [DOI] [PubMed] [Google Scholar]
- 60.Lalzar I, Friedmann Y, Gottlieb Y. Tissue tropism and vertical transmission of Coxiella in Rhipicephalus sanguineus and Rhipicephalus turanicus ticks. Environ Microbiol. 2014;16:3657–3668. doi: 10.1111/1462-2920.12455. [DOI] [PubMed] [Google Scholar]
- 61.Graham JG, MacDonald LJ, Hussain SK, et al. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol. 2013;15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calverley M, Erickson S, Read AJ, Harmsen AG. Resident alveolar macrophages are susceptible to and permissive of Coxiella burnetii infection. PLOS ONE. 2012;7:e51941. doi: 10.1371/journal.pone.0051941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lepidi H, Houpikian P, Liang Z, Raoult D. Cardiac valves in patients with Q fever endocarditis: microbiological, molecular, and histologic studies. J Infect Dis. 2003;187:1097–1106. doi: 10.1086/368219. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez J, Souriau A, Buendia AJ, et al. Experimental Coxiella burnetii infection in pregnant goats: a histopathological and immunohistochemical study. J Comp Pathol. 2006;135:108–115. doi: 10.1016/j.jcpa.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 65.Hellinga JR, Garduno RA, Kormish JD, et al. Identification of vacuoles containing extraintestinal differentiated forms of Legionella pneumophila in colonized Caenorhabditis elegans soil nematodes. Microbiologyopen. 2015;4:660–681. doi: 10.1002/mbo3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brassinga AK, Sifri CD. The Caenorhabditis elegans model of Legionella infection. Methods Mol Biol. 2013;954:439–461. doi: 10.1007/978-1-62703-161-5_27. [DOI] [PubMed] [Google Scholar]
- 67.Fares H, Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Irazoqui JE, Troemel ER, Feinbaum RL, et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLOS Pathog. 2010;6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seervi M, Xue D. Mitochondrial cell death pathways in Caenorhabiditis elegans. Curr Top Dev Biol. 2015;114:43–65. doi: 10.1016/bs.ctdb.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 70.Omsland A, Cockrell DC, Fischer ER, Heinzen RA. Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. J Bacteriol. 2008;190:3203–3212. doi: 10.1128/JB.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Omsland A, Cockrell DC, Howe D, et al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A. 2009;106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DiLoreto R, Murphy CT. The cell biology of aging. Mol Biol Cell. 2015;26:4524–4531. doi: 10.1091/mbc.E14-06-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Akira S, Takeda K. Toll-like receptor signalling. Nature Reviews Immunology. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 75.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 76.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 77.Anderson KV, Schneider DS, Morisato D, et al. Extracellular morphogens in Drosophila embryonic dorsal-ventral patterning. Cold Spring Harb Symp Quant Biol. 1992;57:409–417. doi: 10.1101/sqb.1992.057.01.046. [DOI] [PubMed] [Google Scholar]
- 78.Kanzok SM, Hoa NT, Bonizzoni M, et al. Origin of Toll-like receptor-mediated innate immunity. J Mol Evol. 2004;58:442–448. doi: 10.1007/s00239-003-2565-8. [DOI] [PubMed] [Google Scholar]
- 79.Couillault C, Pujol N, Reboul J, et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 80.Liberati NT, Fitzgerald KA, Kim DH, et al. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim DH, Feinbaum R, Alloing G, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 82.Conti F, Boucherit N, Baldassarre V, et al. Coxiella burnetii lipo-polysaccharide blocks p38alpha-MAPK activation through the disruption of TLR-2 and TLR-4 association. Front Cell Infect Microbiol. 2014;4:182. doi: 10.3389/fcimb.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boucherit N, Barry AO, Mottola G, et al. Effects of Coxiella burnetii on MAPKinases phosphorylation. FEMS Immunol Med Microbiol. 2012;64:101–103. doi: 10.1111/j.1574-695X.2011.00852.x. [DOI] [PubMed] [Google Scholar]
- 84.Barry AO, Boucherit N, Mottola G, et al. Impaired stimulation of p38alpha-MAPK/Vps41-HOPS by LPS from pathogenic Coxiella burnetii prevents trafficking to microbicidal phagolysosomes. Cell Host Microbe. 2012;12:751–763. doi: 10.1016/j.chom.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 85.Rosales EM, Aguilera MO, Salinas RP, et al. Cortactin is involved in the entry of Coxiella burnetii into non-phagocytic cells. PLOS ONE. 2012;7:e39348. doi: 10.1371/journal.pone.0039348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lifshitz Z, Burstein D, Schwartz K, et al. Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen-activated protein kinase pathway. Infect Immun. 2014;82:3740–3752. doi: 10.1128/IAI.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphy CT, McCarroll SA, Bargmann CI, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 88.Garsin DA, Villanueva JM, Begun J, et al. Long-lived C elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 89.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang HC, Paek J, Kim DH. Natural polymorphisms in C elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature. 2011;480:525–529. doi: 10.1038/nature10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hodgkin J, Kuwabara PE, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol. 2000;10:1615–1618. doi: 10.1016/s0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- 92.Couillault C, Ewbank JJ. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect Immun. 2002;70:4705–4707. doi: 10.1128/IAI.70.8.4705-4707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoflich J, Berninsone P, Gobel C, et al. Loss of srf-3-encoded nucleotide sugar transporter activity in Caenorhabditis elegans alters surface antigenicity and prevents bacterial adherence. J Biol Chem. 2004;279:30440–30448. doi: 10.1074/jbc.M402429200. [DOI] [PubMed] [Google Scholar]
- 94.Cipollo JF, Awad AM, Costello CE, Hirschberg CB. srf-3, a mutant of Caenorhabditis elegans, resistant to bacterial infection and to biofilm binding, is deficient in glycoconjugates. J Biol Chem. 2004;279:52893–52903. doi: 10.1074/jbc.M409557200. [DOI] [PubMed] [Google Scholar]
- 95.Gravato-Nobre MJ, Nicholas HR, Nijland R, et al. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2005;171:1033–1045. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Rourke D, Baban D, Demidova M, et al. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yook K, Hodgkin J. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2007;175:681–697. doi: 10.1534/genetics.106.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Partridge FA, Tearle AW, Gravato-Nobre MJ, et al. The C elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev Biol. 2008;317:549–559. doi: 10.1016/j.ydbio.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 99.Parsons LM, Mizanur RM, Jankowska E, et al. Caenorhabditis elegans bacterial pathogen resistant bus-4 mutants produce altered mucins. PLOS ONE. 2014;9:e107250. doi: 10.1371/journal.pone.0107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Palaima E, Leymarie N, Stroud D, et al. The Caenorhabditis elegans bus-2 mutant reveals a new class of O-glycans affecting bacterial resistance. J Biol Chem. 2010;285:17662–17672. doi: 10.1074/jbc.M109.065433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Soltysiak KA, van Schaik EJ, Samuel JE. Surfactant protein D binds to Coxiella burnetii and results in a decrease in interactions with murine alveolar macrophages. PLOS ONE. 2015;10:e0136699. doi: 10.1371/journal.pone.0136699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zamboni DS, Campos MA, Torrecilhas AC, et al. Stimulation of toll-like receptor 2 by Coxiella burnetii is required for macrophage production of pro-inflammatory cytokines and resistance to infection. J Biol Chem. 2004;279:54405–54415. doi: 10.1074/jbc.M410340200. [DOI] [PubMed] [Google Scholar]
- 103.Meghari S, Honstettre A, Lepidi H, et al. TLR2 is necessary to inflammatory response in Coxiella burnetii infection. Ann N Y Acad Sci. 2005;1063:161–166. doi: 10.1196/annals.1355.025. [DOI] [PubMed] [Google Scholar]
- 104.Ochoa-Reparaz J, Sentissi J, Trunkle T, et al. Attenuated Coxiella burnetii phase II causes a febrile response in gamma interferon knockout and Toll-like receptor 2 knockout mice and protects against reinfection. Infect Immun. 2007;75:5845–5858. doi: 10.1128/IAI.00901-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ammerdorffer A, Roest HI, Dinkla A, et al. The effect of C burnetii infection on the cytokine response of PBMCs from pregnant goats. PLOS ONE. 2014;9:e109283. doi: 10.1371/journal.pone.0109283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Honstettre A, Ghigo E, Moynault A, et al. Lipopolysaccharide from Coxiella burnetii is involved in bacterial phagocytosis, filamentous actin reorganization, and inflammatory responses through Toll-like receptor 4. J Immunol. 2004;172:3695–3703. doi: 10.4049/jimmunol.172.6.3695. [DOI] [PubMed] [Google Scholar]
- 107.Kubes M, Kuzmova Z, Gajdosova E, et al. Induction of tumor necrosis factor alpha in murine macrophages with various strains of Coxiella burnetii and their lipopolysaccharides. Acta Virol. 2006;50:93–99. [PubMed] [Google Scholar]
- 108.Bradley WP, Boyer MA, Nguyen HT, et al. Primary role for TLR-driven TNF rather than cytosolic immune detection in restricting Coxiella burnetii phase II replication within mouse macrophages. Infect Immun. 2016;84:998–1015. doi: 10.1128/IAI.01536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moos A, Hackstadt T. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect Immun. 1987;55:1144–1150. doi: 10.1128/iai.55.5.1144-1150.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kersh GJ, Oliver LD, Self JS, et al. Virulence of pathogenic Coxiella burnetii strains after growth in the absence of host cells. Vector Borne Zoonotic Dis. 2011;11:1433–1438. doi: 10.1089/vbz.2011.0670. [DOI] [PubMed] [Google Scholar]
- 111.Hackstadt T. Steric hindrance of antibody binding to surface proteins of Coxiella burnetti by phase I lipopolysaccharide. Infect Immun. 1988;56:802–807. doi: 10.1128/iai.56.4.802-807.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vishwanath S, Hackstadt T. Lipopolysaccharide phase variation determines the complement-mediated serum susceptibility of Coxiella burnetii. Infect Immun. 1988;56:40–44. doi: 10.1128/iai.56.1.40-44.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shannon JG, Howe D, Heinzen RA. Virulent Coxiella burnetii does not activate human dendritic cells: role of lipopoly-saccharide as a shielding molecule. Proc Natl Acad Sci USA. 2005;102:8722–8727. doi: 10.1073/pnas.0501863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nicholas HR, Hodgkin J. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr Biol. 2004;14:1256–1261. doi: 10.1016/j.cub.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 115.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]