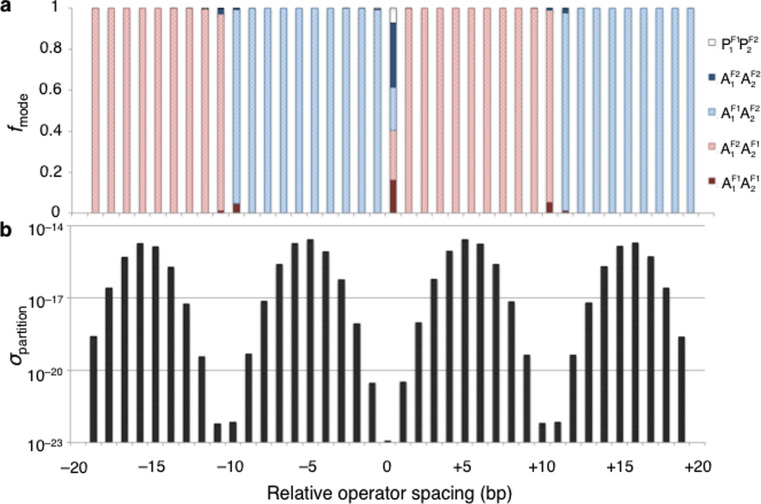
Fig. 5.
Profiles, as a function of operator spacing, of (a) the fraction f mode of 182-bp, Lac repressor-bound DNA minicircles in each of the energetically favored modes of loop attachment to the V-shaped protein assembly and (b) the relative ease of partitioning the DNA into two domains at these sites. Operator spacing is expressed in terms of the displacement of the recognition sequences, in base-pair steps, relative to the even positioning at 0 associated with the formation of equally sized loops having the 92-bp center-to-center spacing found in the Escherichia coli lac operon. That is, one of the protein-anchored loops is altered in size by the specified difference in spacing and the other by its negative. The ease of DNA partitioning σ partition is given by the sum of the Boltzmann factors of the elastic energies of the modeled Lac repressor-bound minicircles at each spacing. Note the much lower chances of dividing the minicircle in half compared to partitioning it at other sites