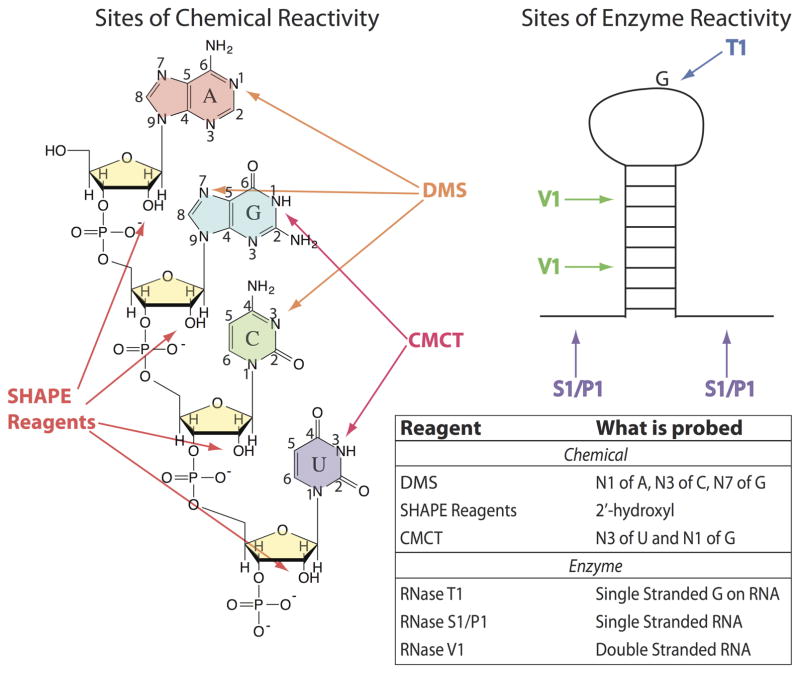
Figure 6.
RNA modifications by DMS (dimethyl sulfate), SHAPE reagents (Selective 2′-hydroxyl acylation analyzed by primer extension), and CMCT (1-cyclohexyl-(2-morpholinoehyl)carbodiimide metho-p-toluene). SHAPE reagents modify the 2′-hydroxyl on the sugar of all four nucleobases. SHAPE reagents include 1M7 (1-methyl-7-nitroisatoic anhydride), NMIA (N-methylisotoic anhydride), and NAI (2-methylnicotinic acid imidazolide). DMS modifies the N1 of A and the N3 of C as well as the N7 of G. CMCT modifies N3 of U and N1 of G. The chemical modifications (except N7 of G) can be detected immediately by RT followed by gel electrophoresis or high-throughput sequencing, and the enzymatic cleavages, which modify single- and double-stranded RNA, can be read out through gel electrophoresis or high-throughput sequencing.