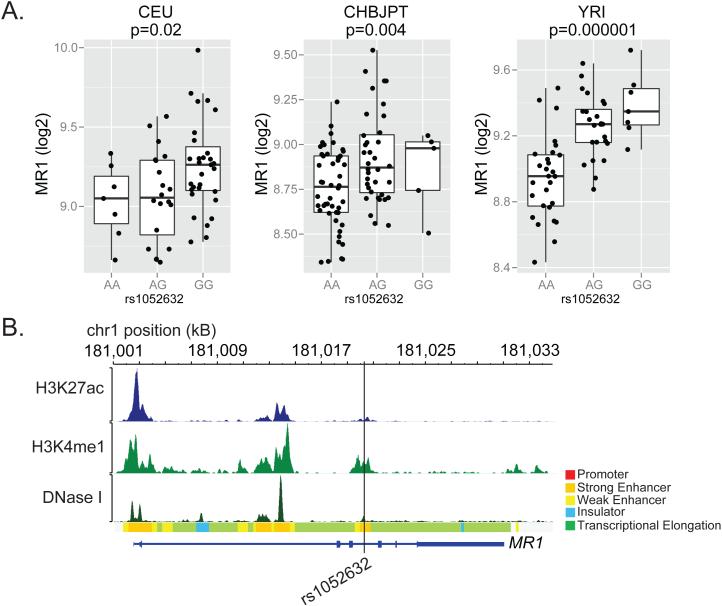
Figure 3. rs1052632 is associated with MR1 expression and is located within a transcriptional enhancer.
(A) Genome-wide transcriptional analysis of lymphoblastoid cell lines derived from HapMap participants42 was merged with subject-specific MR1 genotypes downloaded from (http://hapmap.ncbi.nlm.nih.gov/downloads/genotypes/2007-01/fwd_strand/non-redundant/). Generalized linear regression was used to determine the association between rs105632 genotype and MR1 expression in Caucasians of European descent (CEU, n=60), Chinese in Beijing or Japanese in Tokyo (CHBJPT, n=90), or Yorubans in Nigeria (YRI, n=60). The boxplots show log2 transformed fluorescence intensity of MR1 expression as measured by microarray stratified by rs1052632 genotype. (B) Functional annotation for rs1052632 within normal human epidermal keratinocytes (NHEK) was downloaded and visualized using the RoadMap Epigenomics web browser (http://epigenomegateway.wustl.edu/browser/). DNase I represents DNase hypersensitive sites, H3K27ac represents acetylation of lysine 27 on histone H3, and H3K4me1 represents methylation of lysine 4 on histone H3. The Chromatin State is derived from a Hidden Markov Model using ChIP-Seq data for nine parameters measured in nine human cell types through the ENCODE Project 43.