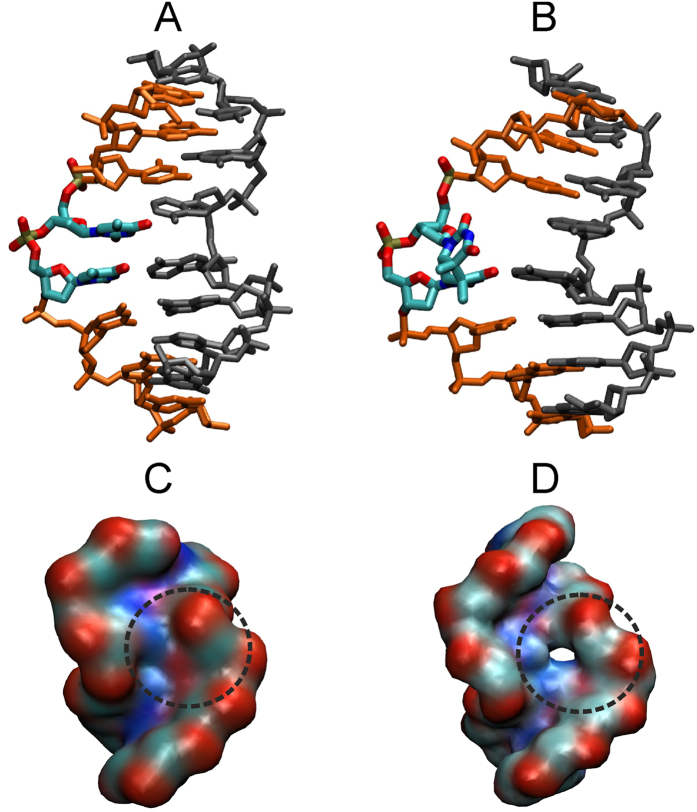
Figure 3. Free energy minima for the central damaged or undamaged DNA obtained during free energy simulations of DNA in near bound conformation.
(A) The free energy minimum obtained for the regular TT case near 30° along the flipping reaction coordinate indicates a base-paired central arrangement (strands in orange and grey, central TT nucleotides in atom color code). (B) The free energy minimum obtained for the CPD damage is located at 50° along the reaction coordinate and indicates a structure that is partially looped out into the major groove, at same time creating sterically accessible space on the minor groove surface of the DNA. (C) View into the minor groove of the free energy minimum obtained for regular DNA globally restraint to the repair enzyme bound form (atom-color coded surface representation). (D) Same as (C) but for the CPD damaged case clearly indicating sterically accessible space at the CPD damage site. The protein binding region (with a central Pro-residue fitting into the sterical accessible space).