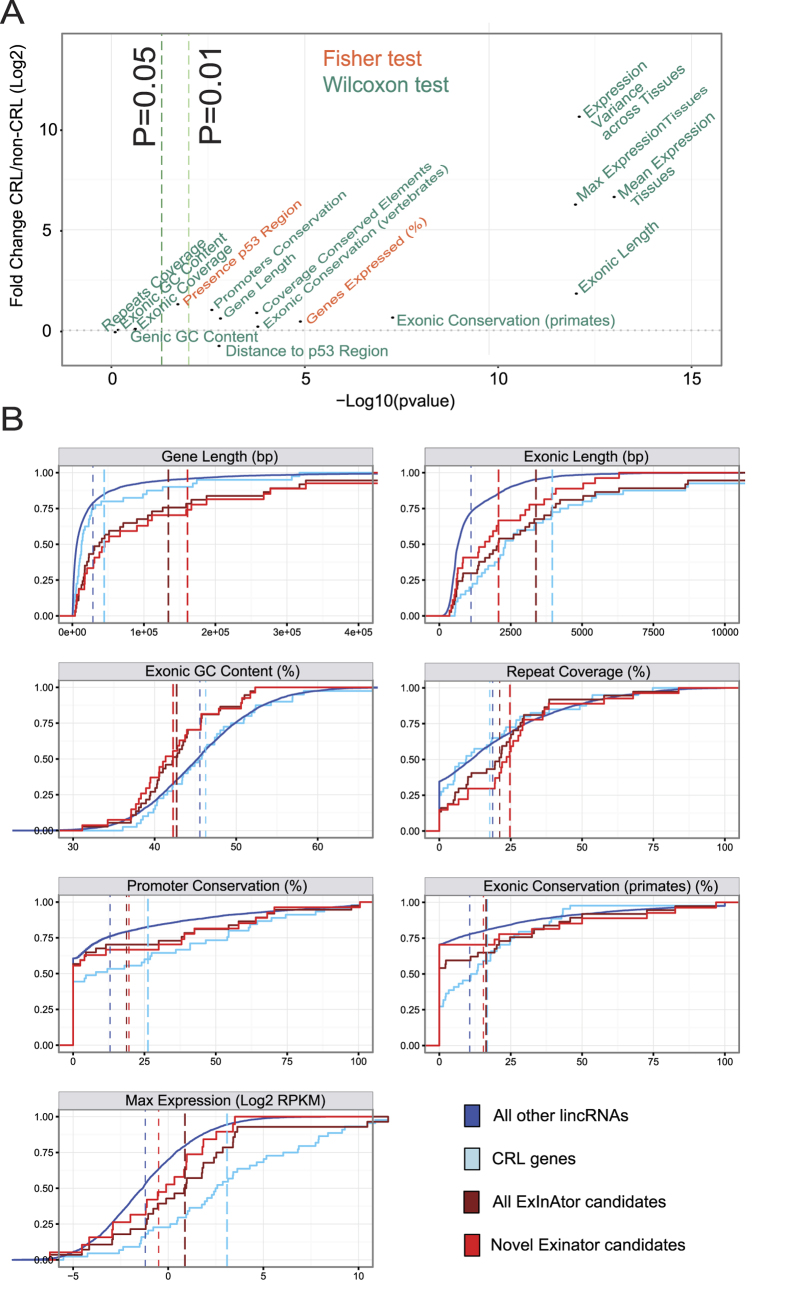
Figure 5. Features distinguishing cancer lncRNAs are also found in ExInAtor candidates.
(A) Identification of cancer lncRNA features by comparing literature-curated Cancer Related LncRNA (CRL) genes to others. Dots represent 16 features that were compared in CRL and non-CRL genes. y axis shows the log2 fold difference of CRL vs non-CRL means for the values of the given feature. x axis represents the P value obtained from the statistical test applied when comparing CRL and non-CRL. Features are coloured depending on whether they are discrete features, analysed by Fisher Exact test, or continuous features analysed by Wilcoxon test. (B) Cancer lncRNA features in ExInAtor-predicted driver genes. Shown are cumulative distributions for seven selected features. Dashed vertical lines indicate the mean value of each group. Genes are grouped by: literature-reported cancer CRL lncRNAs, all ExInAtor candidates (both CRL and not) (“All candidates”), only novel ExInAtor candidates that are not included in CRL (“novel candidates”), and non-CRL non-candidates, being all other GENCODE lincRNAs. Candidates here were defined at a Q ≤ 0.2. Groups significantly different from the latter at a threshold of P = 0.05 (Wilcoxon test) are represented by a thick line.