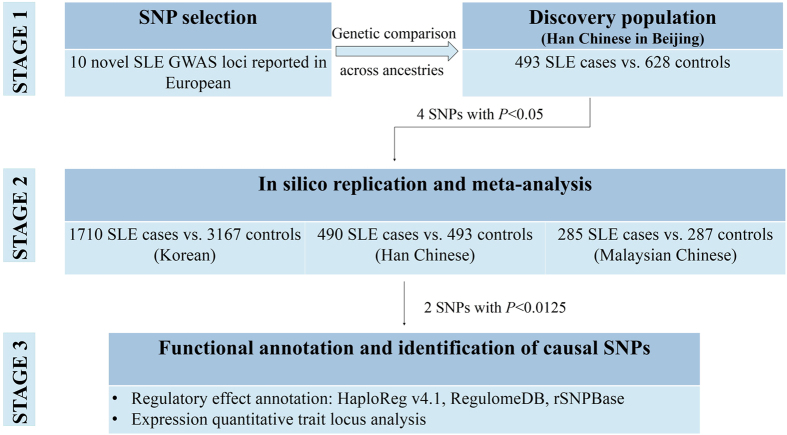
Figure 1. Workflow of our study design.
The study was designed in three stages. First, we genotyped the 10 novel genome wide associated loci with European SLE patients in a Han Chinese cohort in Beijing and compared the genetic similarities and differences between the two ancestries. Four out of 10 loci were identified as significant (P < 0.1). Second, we performed independent replications of these 4 loci in three cohorts from Korean, Han Chinese and Malaysian Chinese. Consistent associations have been identified in our discovery and replication cohorts for two loci (i.e., IL12A and TCF7). Third, by integrating different layers of functional data, we identified the most likely functional SNPs for these two loci. Abbreviations: GWAS: genome-wide association study; SLE: systemic lupus erythematosus; SNP: single nucleotide polymorphism.