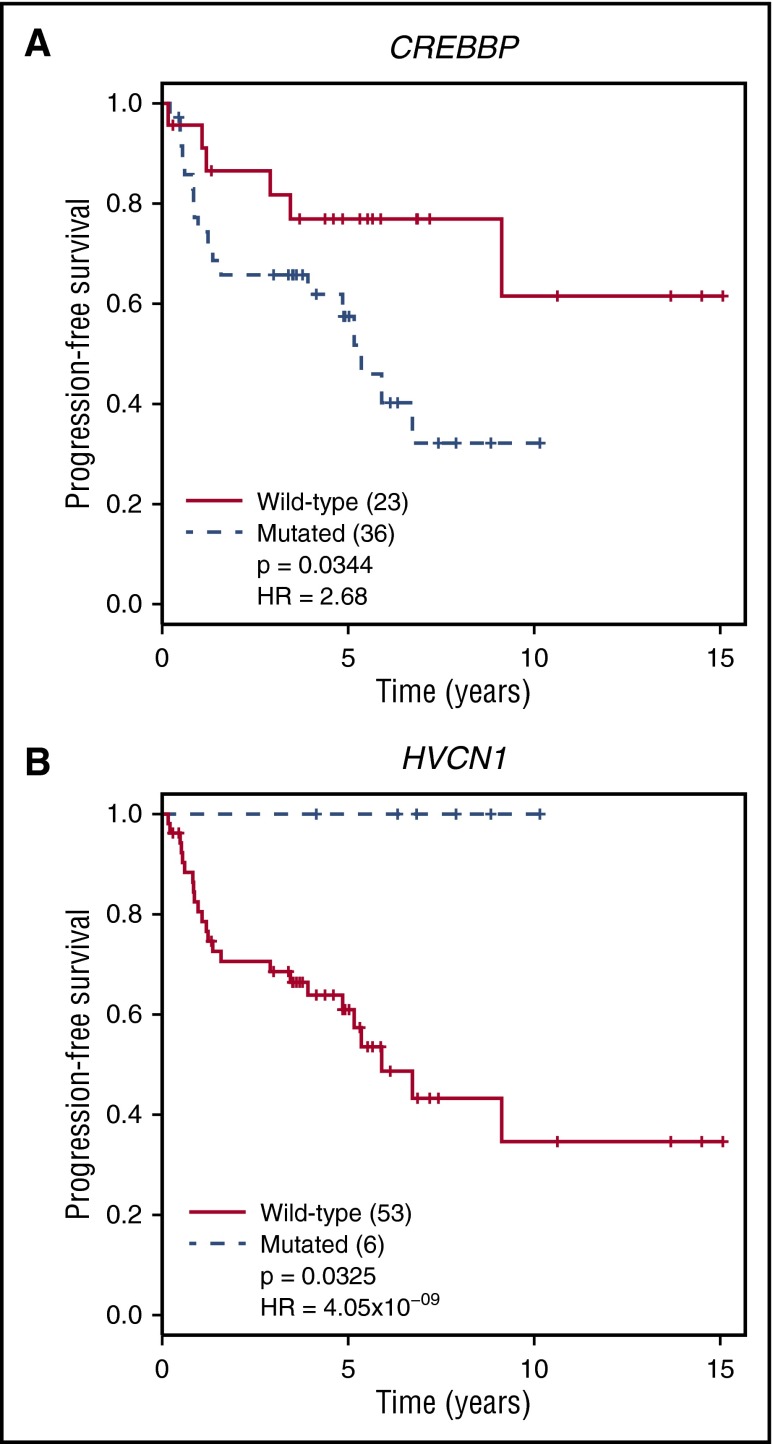
Figure 6.
Mutations affecting PFS in treated patients with FL. Treatment-naive patients who received treatment within 1 year of diagnosis and sample collection (N = 59) were stratified by the presence or absence of coding or splice site mutations in SMGs, with a minimum of 5 mutations in this subset of patients (supplemental Table 6). Only groups showing significantly different survival are shown. (A) PFS was worse for patients harboring CREBBP mutations (P = .034; q = 0.884 after Benjamini-Hochberg correction for multiple hypothesis testing). (B) In contrast, patients with HVCN1 mutations had better PFS than those with wild-type HVCN1 (P = .033; q = 0.740).