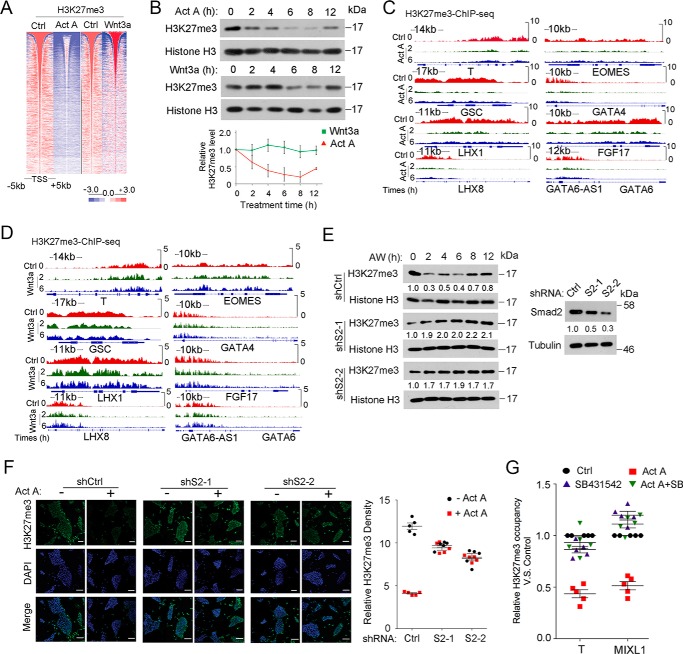
FIGURE 5.
Activin/Smad2 signaling is critical for H3K27me3 reduction. A, heat map of H3K27me3 and H3K4me3 intensity centered at all of the detected peaks after H1 cells were treated with 25 ng/ml activin A or 25 ng/ml Wnt3a for 2 h before they were harvested for the ChIP assay followed by high throughput sequencing against H3K27me3 and H3K4me3 antibody. B, H1 cells treated with 25 ng/ml activin A or 25 ng/ml Wnt3a for the indicated times before they were harvested for histone extraction followed by anti-H3K27me3 immunoblotting. H3K27me3 bands were quantified, and the relative levels were shown after normalization to histone H3. A Friedman test was performed, and the statistical data are shown as mean ± S.E. (error bars) (n = 9, including 3 biological replicates and 3 technical replicates). C and D, IGV showed examples of H3K27me3 enrichment at 2 and 6 h on specific ME genes as revealed by H3K27me3 ChIP-seq upon activin A (C) or Wnt3a treatment (D). E, Smad2 knockdown H1 cells were treated with 25 ng/ml AW in B27 medium for the indicated times before they were harvested for histone extraction followed by anti-H3K27me3 immunoblotting. Nonspecific target shRNA was used as control. Knockdown efficiency of Smad2 was shown in the left panel. Smad2 and H3K27me3 bands were quantified. F, Smad2 knockdown H1 cells treated with 25 ng/ml activin A for 2 h before they were harvested for anti-H3K27me3 immunofluorescence. H3K27me3 densities were quantified with ImageJ. The nucleus was counterstained with DAPI. Scale bar, 100 μm. G, H1 cells treated with 25 ng/ml activin A, 10 μm SB431542, or both for 2 h before they were harvested for anti-H3K27me3 ChIP followed by qPCR. The amplified promoter regions for ChIP-qPCR were as follows: T at about 1 kb upstream of TSS; MIXL1 at about 0.8 kb upstream of the TSS. The statistical data are shown as mean ± S.E. (F, n = 5, including 5 biological replicates; G, n = 15, including 5 biological replicates and 3 technical replicates). A two-way ANOVA test was used in F and G.