Fig. 3.
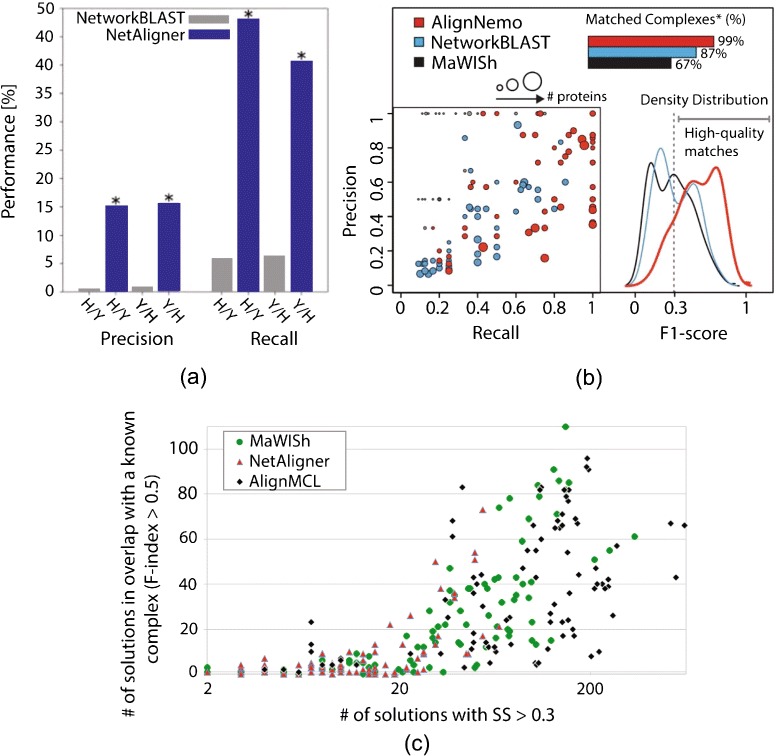
Illustration of the performance of different local network aligners. The figure compares a NetworkBLAST and NetAligner, b AlignNemo, NetworkBLAST, and MaWISh, and c MaWISh, NetAligner, and AlignMCL, with respect to precision, recall, or F-score of “aligned network module–known protein complex” matches. In a, precision and recall are shown when aligning networks of human (H) and yeast (Y), where the order of networks (i.e., H/Y versus Y/H) plays a role. The statistical significance of the performance difference between the two methods is indicated with an asterisk. In b, results are shown only for conserved network modules with more than six nodes, when aligning yeast and fly networks. In the lower left, precision versus recall is shown for each conserved module, represented as a circle whose radius is proportional to the size of the module. In the lower right, F-score distributions are shown for each method. Finally, at the top right, percentages are shown to quantify how well the given method’s conserved modules match known protein complexes. In c, each point represents an alignment. The position of a point on y-axis is determined by the number of modules (or solutions) conserved under the given alignment that match known protein complexes with F-score (i.e., F-index) above 0.5. The position of a point on x-axis is determined by the number of modules conserved under the given alignment that have semantic similarity (SS) scores above 0.3; semantic similarity of a conserved module quantifies functional homogeneity of proteins within the module. a, b, and c are adopted from [52, 54] and [53], respectively
