ABSTRACT
mRNA (mRNA) transport focuses the expression of encoded proteins to specific regions within cells providing them with the means to assume specific functions and even identities. BicD and the mRNA binding protein Egl interact with the microtubule motor dynein to localize mRNAs in Drosophila. Because relatively few mRNA cargos were known, we isolated and identified Egl::GFP associated mRNAs. The top candidates were validated by qPCR, in situ hybridization and genetically by showing that their localization requires BicD. In young embryos these Egl target mRNAs are preferentially localized apically, between the plasma membrane and the blastoderm nuclei, but also in the pole plasm at the posterior pole. Egl targets expressed in the ovary were mostly enriched in the oocyte and some were apically localized in follicle cells. The identification of a large group of novel mRNAs associated with BicD/Egl points to several novel developmental and physiological functions of this dynein dependent localization machinery. The verified dataset also allowed us to develop a tool that predicts conserved A'-form-like stem loops that serve as localization elements in 3′UTRs.
KEYWORDS: Bicaudal-d, Drosophila development, egalitarian, mRNA localization, microtubule transport
Introduction
mRNA transport is an important mechanism for focusing protein expression to specific regions of cells, to individual cells in a group of interconnected cells, or even to regions of an organism in cases where its nuclei share a common cytoplasm. mRNA localization plays important functions wherever spatial restriction of protein expression is needed and the importance of mRNA localization for normal development and physiology is underlined by the fact that this mechanism is found form bacteria to humans.1 Drosophila oocytes and embryos are well-established systems to study mRNA localization. Oocyte determination depends on mRNA localization and the spatial localization of bicoid (bcd), oskar (osk) and gurken (grk) transcripts determines the embryonic anterior-posterior axis as well as the dorsal-ventral axis already during oogenesis.2 During the syncytial blastoderm stage of embryonic development pair-rule transcripts, such as fushi tarazu (ftz), even-skipped (eve) and hairy (h) become localized via microtubule (MT) minus-end directed transport to the apical cytoplasm, above the layer of nuclei. Their local translation in the apical cytoplasm close to the nuclei allows the local synthesis of the transcription factors encoded by them. This is an efficient way to produce the proteins where they are needed and to restrict diffusion to the surrounding nuclei. Other transcripts that are targeted apically to the nuclei are the mRNA encoding the signaling molecule Wingless (Wg), which is subsequently translated and secreted apically, and the inscuteable (insc) mRNA that becomes apically localized in neuroblasts.3,4,5 Altogether, 71% of the genes expressed during Drosophila embryonic development show discernable subcellular mRNA localization patterns.6
In Drosophila, mRNA localization involves a machinery composed of Bicaudal-D (BicD) and Egalitarian (Egl), which interact with dynein/dynactin microtubule motors on the one hand and with the mRNA cargo on the other (reviewed in1). In this way the mRNA cargo is targeted to distinct cellular compartments. While we have learned much about the genes and proteins involved in mRNA transport and localization, until recently we knew only a small number of mRNAs that use the BicD/Egl localization machinery for their transport. Here we describe many novel mRNAs localized by BicD/Egl. The results for the top candidates were validated in independent experiments and in several ways. Aside from quantifying their enrichment during immunopurification with qPCR, we also included genetic analyses combined with in situ mRNA localization experiments to confirm that these mRNAs indeed require BicD to reach their target regions. This approach also revealed novel cellular compartments that are targeted by the BicD/Egl transport machinery in embryos and ovaries. Finally we used the resource we established to develop a search algorithm to identify putative A'-form-like localizing elements in 3′UTRs. Testing two of the newly identified putative localization sequences revealed that the program successfully identified novel localizing signals in the 3′UTR of novel Egl targets.
Results
RIPseq uncovers novel putative Egl targets
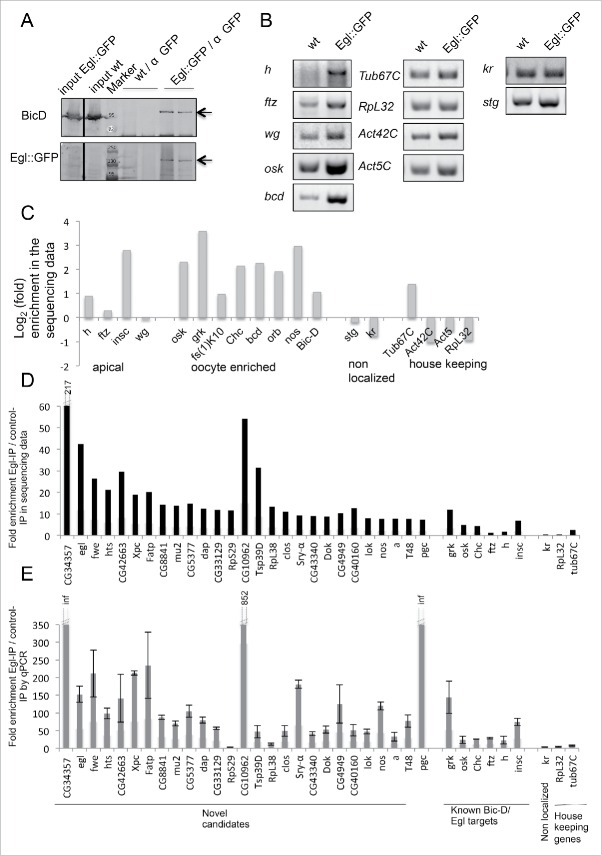
Anti-GFP antibodies were used to immunoprecipitate Egl-associated mRNAs from cytoplasmic extracts prepared from 0–8 hours old Drosophila embryos expressing a functional Egl::GFP fusion protein.7 As control for unspecific binding we used extracts from wild type embryos lacking the GFP epitope. Immunoprecipitation (IP) conditions were optimized by analyzing the enrichment of known BicD/Egl mRNA targets in the experimental IP compared to the control IP (Fig. 1A, B). This procedure efficiently co-immunoprecipitated Egl::GFP, BicD and endogenous Egl (Fig. 1A and data not shown). Because BicD forms dimers,8,9 this likely indicates that a multimeric Egl::GFP/BicD/BicD/Egl complex was pulled down. The apically localizing mRNAs h, ftz and wg4,10 were strongly enriched in the Egl::GFP precipitates compared to the controls (Fig. 1B). The abundance of 2 mRNAs that get localized through BicD/Egl during oogenesis (osk, bcd) was also elevated in the embryonic Egl::GFP IP. As negative controls we tested the known non-localized transcripts from Krüppel (Kr) and string (stg), which are not recruited to the BicD/Egl localization machinery in embryos10 and we included the house keeping genes Tub67C, RpL32, Act5C and Act42C (Fig. 1B). While we observed residual amplification of all transcripts in the control IPs (wt; probably due to minor amounts of unspecific binding to the beads), only localized mRNAs showed a clear enrichment in the Egl::GFP IP (Fig. 1B). Enrichment was observed when using the same amount of pulled down total RNA (Fig. 1B) and the same amount of initial embryonic material, respectively (Fig. S1).
Figure 1.
RipSeq protocol retains BicD/Egl complex stability and enriches for known target mRNAs. (A-B) Extracts from Egl::GFP expressing embryos and wt control extracts were subjected to IP with anti-GFP antibodies. Western blotting tested for the presence of the BicD and Egl::GFP (A). Semiquantitative RT-PCR tested for the enrichment of known BicD/Egl mRNA targets and for the lack of enrichment of non-localizing mRNAs and house keeping mRNAs (B). (C) Rip-Seq enrichment of mRNAs from the test set. (D-E) Most top candidate BicD/Egl targets identified by Rip-Seq (D) could be validated by RT-qPCR analysis in an independent IP experiment (E). Error bars represent +/− SD of 2 independent IPs. Note that fold enrichment values of the 2 different experiments cannot be compared directly because they are calculated using different formulas (see methods). Nevertheless, enrichments in the Egl::GFP IPs relative to mock IPs is reproducible.
Immunoprecipitations were performed with 2 independent biological samples of Egl::GFP and controls. Biological replicates were pooled for Illumina sequencing and enrichment factors and P-values were determined and compiled (Table S1). Many of the known BicD/Egl mRNA cargoes such us osk, bcd, insc, nos, orb and Chc mRNAs were found to be highly enriched in the Egl::GFP IPs and they ranked in the top 140 enriched mRNAs, while our non-localizing mRNAs were not in this group (Fig. 1C; Table S1). Therefore, the preparative purification scheme effectively enriched for Egl mRNA targets. Using the known target osk mRNA as a cut off, a list of the top 50 mRNAs with the highest enrichment scores was produced. Table 1 lists the embryonic expression data for these genes and Table 2 their oogenesis expression data. These predicted targets were selected for further analysis. Interestingly, many top candidate mRNAs show higher enrichment values than the already described Egl targets (Fig. 1D). Because the known BicD/Egl targets had been identified by genetic and not by biochemical experiments and because many of them perform their function primarily in the ovarian germ line, it is not surprising that we identified mRNAs that are more enriched than the already known ones. To our knowledge this was the first biochemical approach taken to identify targets of this machinery and it revealed numerous novel mRNAs that seem to attach significantly and specifically to Egl:GFP.
Table 1.
Localization patterns of Egl::GFP target mRNA candidates in embryonic stages 1–5.
| Gene-ID | pVal | Fold change | Embryo expression (St. 1 to 5, our data) | Embryo expression (St. 1 to 5, published-data) | Maternal expression |
|---|---|---|---|---|---|
| CG34357 | 1.08E-06 | 217.53 | NE | NA | |
| egl | 2.10E-05 | 42.33 | Ubiquitous, maternal | Ubiquitous, degraded by st.4 (Mach and Lehmann, 1997) (BDPG) | ✓ |
| snmRNA:331 | 1.60E-04 | 24.87 | NA | NA | |
| CG6151 | 1.60E-04 | 26.38 | Not expressed | Ubiquitous- weak (BDPG) | ✓ |
| hts | 3.03E-04 | 21.1 | NA | Anterior gradient (Ding et al., 1993) (Yue and Spradling, 1992) | ✓ |
| CG42663 = | |||||
| CG12488 | 4.34E-04 | 29.6 | Ubiquitous-weak | Not expressed (BDPG) | |
| mus210 = Xpc | 4.63E-04 | 19 | NA | Anterior gradient. apical enrichment (BDPG) | ✓ |
| Fatp = CG7400 | 9.80E-04 | 20.15 | Ubiquitous-weak | Weak anterior gradient (BDPG) | ✓ |
| CG8841 | 1.37E-03 | 14.4 | Gap expression pattern- apical and pole cell enrichment | Maternal (BDPG) | ✓ |
| mu2 | 1.59E-03 | 13.9 | NA | Anterior gradient (Kasravi et al., 1999) | ✓ |
| CG5377 | 2.00E-03 | 14.8 | Ubiquitous-weak | Ubiquitous, early degraded (BDPG) | ✓ |
| dap | 2.28E-03 | 12.4 | NA | Ubiquitous. Pole plasm- pole cell enrichment (BDPG) | ✓ |
| CG33129 | 2.50E-03 | 11.9 | Pole cell enrichment-weak | Ubiquitous (BDPG), pole cell enrichment (Fly-Fish) | ✓ |
| RpS29 | 2.60E-03 | 11.7 | Ubiquitous-strong | Ubiquitous (BDPG) | ✓ |
| CG10962 | 2.62E-03 | 54.1 | Ubiquitous-weak | Ubiquitous, apical exclusion St. 4–5 (Fly-Fish) | ✓ |
| grk | 3.26E-03 | 12 | NA | Ubiquitous-weak, degraded completely st.5 (Fly-Fish) | ✓ |
| Tsp39D | 3.49E-03 | 31.3 | Ubiquitous-weak | NA | |
| RpL38 | 3.73E-03 | 13.4 | Ubiquitous | NA | |
| Acf1 | 3.94E-03 | 10.6 | Apical enrichment in cycles 13–14 | Ubiquitous, degradation of maternal transcripts (Fly-Fish, BDPG) | ✓ |
| CG42600 = clos | 3.96E-03 | 11.1 | Basal localization | NA | |
| Sry-α | 5.87E-03 | 9.4 | Perinuclear- apical enrichment-strong | Perinuclear St. 1–3, apical enrichment, St. 4–5 (Fly-Fish). Ubiquitous (BDPG) | ✓ |
| CG43340 = | |||||
| CG30492 | 6.37E-03 | 9.1 | Basal localization | Ubiquitous, degradation maternal transcripts (Fly-Fish) | ✓ |
| Dok | 6.85E-03 | 8.9 | Apical and pole cell enrichment (weak) | Ubiquitos, perinuclear. St. 4–5 apical and pole cell enrichment (Biswas et al., 2006, Fly-Fish, BDPG) | ✓ |
| CG4949 | 8.39E-03 | 10.3 | NA | Ubiquitous, St. 1–3, St. 4–5 apical enrichment (Fly-Fish ). Ubiquitous faint, St. 1–3, St. 4–5 apical and pole cell enrichment (BDPG) | ✓ |
| CG40160 | 1.02E-02 | 12.6 | Weak ubiquitous | Weak ubiquitous (Fly-Fish) | ✓ |
| lok | 1.03E-02 | 8 | Gap-expression-anterior gradient.St 4–5 segmented pattern. Apical, pole plasm and pole cell enrichment | Gap-expression-anterior gradient. St 4–5 segmented pattern, apical and pole plasm, pole cell enrichment (Fly-Fish, Oishi et al., 1998, BDPG) | ✓ |
| nos | 1.04E-02 | 7.8 | Pole plasm and pole cell enrichment, maternal | Pole plasm and pole cell enrichment. (Wang and Lehmann, 1991) | ✓ |
| a | 1.05E-02 | 7.9 | Apical-anterior gradient | First ubiquitous and then gap-expression anterior gradient (Liu and Lengyel, 2000) | ✓ |
| T48 | 1.22E-02 | 7.8 | Apical- ventral enrichment | Ventral enrichment (Kölsch et al.2007) | |
| pgc | 1.41E-02 | 7.4 | Pole plasm and pole cell enrichment, maternal | Pole plasm and pole cell enrichment, perinuclear (Martinho et al., 2004, BDPG, Fly-Fish) | ✓ |
| Rab9 | 1.44E-02 | 9.4 | Basal localization | Maternal (BDPG) | ✓ |
| CG12945 | 1.60E-02 | 7.7 | Basal localization-faint staining | Maternal, rapidly degraded (BDPG) | ✓ |
| insc | 1.78E-02 | 6.8 | NA | NE (BDPG) | |
| pbl | 1.81E-02 | 6.7 | NA | Pole plasm and pole cells (Prokopenko et al., 2000, Fly-Fish) | ✓ |
| didum | 1.95E-02 | 6.6 | Ubiquitous | Ubiquitous (Bonafe and Sellers, 1998) | ✓ |
| RpS15 | 2.03E-02 | 6.3 | Ubiquitous-strong | Ubiquitous (BDPG) | ✓ |
| tefu | 2.11E-02 | 7.7 | Ubiquitous -weak | Basal, apical and pole cell exclusion (Fly-Fish) | ✓ |
| snoRNA:U3:9B | 2.13E-02 | 19.4 | NA | NA | |
| fs(1)N | 2.40E-02 | 6.1 | Ubiquitous | Maternal deposited, degraded by St 4 (BDPG) | ✓ |
| CG17698 | 2.51E-02 | 10.1 | Ubiquitous | St. 1–3 no expression, St. 4–5, faint ubiquitous, yolk nuclei (BDPG) | |
| nmdyn-D6 | 2.74E-02 | 13.4 | Ubiquitous-weak | NA | |
| CG33298 | 2.88E-02 | 6.2 | Ubiquitous-weak | St. 1–3 ubiquitous, hindgut anlage in statu nascendi (BDPG). St.1–3 ubiquitous weak, St. 4–5 subset anterior blastoderm nuclei (Fly-Flish) | ✓ |
| CG6459 | 2.91E-02 | 5.9 | Ubiquitous-weak | St .1–3, ubiquitous-weak, degraded completely at St. 5 (BDPG, Fly-Fish) | ✓ |
| CG5604 | 3.00E-02 | 5.7 | Ubiquitous-weak | No expression (BDPG) | |
| ptr | 3.30E-02 | 5.5 | Ubiquitous-weak | St .1–3, ubiquitous-weak, degraded completely at St. 5 (BDPG, Fly-Fish) | ✓ |
| CG5877 | 3.89E-02 | 6.3 | Ubiquitous-weak | NA | |
| Mitf | 3.96E-02 | 5.7 | Basal enrichment-posterior enrichment | Procephalic ectoderm anlage in statu nascendi (BDPG) | ✓ |
| CG3585 | 3.97E-02 | 5.1 | Basal enrichment-weak | NA | ✓ |
| Uba1 | 4.17E-02 | 4.9 | Maternal, ubiquitous. Apical enrichment-weak | Ubiquitous St.1–5 (BDPG). Not expressed (Fly-Fish) | ✓ |
| osk | 4.29E-02 | 4.9 | Pole plasm and pole cells | Pole plasm and pole cells (Ephrussi et al., 1991) | ✓ |
Top Egl::GFP mRNAs are sorted by pVal. NA: not analyzed; NE: no expression; ✓: these genes show maternal expression. Known Egl targets are grk, nos, insc, osk.
Table 2.
Localization patterns of Egl::GFP target mRNA candidates in ovaries.
| Gene-ID | Ovary expression | DOT (Dresden Ovary table) | Consistent with being transported by BicD/Egl in ovaries and/or embryos |
|---|---|---|---|
| CG34357 | NE | ND | NE |
| egl | Oocyte enrichment (Mach and Lehmann 1997), Apical in follicle cell epithelia | Oocyte enrichment. Apical in follicle cell epithelia | ✓ |
| snmRNA:331 | NA | ND | NA |
| CG6151 | Oocyte enrichment | Oocyte enrichment | ✓ |
| hts | Oocyte enrichment (Yue and Spradling, 1992) | ND | ✓ |
| CG42663 = CG12488 | Oocyte enrichment | ND | ✓ |
| mus210 = Xpc | Oocyte enrichment | Oocyte enrichment | ✓ |
| Fatp = CG7400 | Oocyte enrichment | Oocyte enrichment | ✓ |
| CG8841 | Ubiquitous-strong (oocyte and nc expression equal) | ND | ✓ |
| mu2 | Oocyte enrichment (Kasravi et al., 1999) | ND | ✓ |
| CG5377 | Ubiquitous-very weak | ND | |
| dap | Oocyte enrichment (de Nooij, J.C., et al., 2000) | ND | ✓ |
| CG33129 | Oocyte enrichment- Apical in follicle cell epithelia | Oocyte enrichment- Apical in follicle cell epithelia | ✓ |
| RpS29 | Ubiquitous-strong (stronger in nc than in the oocyte) | ND | |
| CG10962 | Ubiquitous-strong (oocyte and nc expression equal) | Nurse cell nuclei foci | ✓ |
| grk | Oocyte enrichment (Neuman-Silberberg and Schupbach, 1993) | Oocyte enrichment | ✓ |
| Tsp39D | Ubiquitous-very weak | ND | |
| RpL38 | Ubiquitous- strong (stronger in nc than oocyte) | ND | |
| Acf1 | Oocyte enrichment | ND | ✓ |
| CG42600 = clos | Oocyte enrichment | ND | ✓ |
| Sry-α | Ubiquitous-weak | ND | ✓ |
| CG43340 = CG30492 | Oocyte enrichment | ND | ✓ |
| Dok | Oocyte enrichment | Oocyte enrichment | ✓ |
| CG4949 | NA | Oocyte enrichment | ✓ |
| CG40160 | Ubiquitous (stronger in nc than in the oocyte) | Somatic cells follicle cells, no images | ✓ |
| lok | Oocyte enrichment (Oishi et al., 1998) | Oocyte enrichment | ✓ |
| nos | Oocyte enrichment (Wang et al., 1994) | ND | ✓ |
| a | Oocyte enrichment | ND | ✓ |
| T48 | Oocyte enrichment | ND | ✓ |
| pgc | Oocyte enrichment | Oocyte enrichment | ✓ |
| Rab9 | Oocyte enrichment (St.2 to 6) | Oocyte enrichment and/or ubiquitous (pattern not reproducible) | ✓ |
| CG12945 | Ubiquitous-weak (stronger in nc than in the oocyte) | ND | |
| insc | Oocyte enrichment | Oocyte enrichment | ✓ |
| pbl | NA | ND | ✓ |
| didum | Oocyte enrichment (MacIver et al., 1998) | Oocyte enrichment | ✓ |
| RpS15 | Ubiquitous-strong (slightly stronger in nc than oocyte) | ND | |
| tefu | Ubiquitous-weak (oocyte and nc expression equal) | ND | ✓ |
| snoRNA:U3:9B | NA | ND | NA |
| fs(1)N | oocyte enrichment | Oocyte enrichment | ✓ |
| CG17698 | Ubiquitous-strong (slightly stronger in nc than oocyte) | Nuclear foci somatic cells nc and follicle cells | |
| nmdyn-D6 | Ubiquitous-very weak | ND | |
| CG33298 | Ubiquitous-strong (oocyte and nc expression equal) | ND | ✓ |
| CG6459 | Ubiquitous-strong (stronger in nc than in the oocyte in ealry stages). St. 9 more accumulation in the oocyte. | ND | ✓ |
| CG5604 | Oocyte enrichment (St.4 to 6) | ND | ✓ |
| Ptr | Ubiquitous-strong (oocyte and nc expression equal) | ND | ✓ |
| CG5877 | Ubiquitous-strong (stronger in nc than oocyte) | ND | |
| Mitf | Ubiquitous (oocyte and nc expression equal) | ND | ✓ |
| CG3585 | Oocyte enrichment (St.4 to 6) | ND | ✓ |
| Uba1 | Oocyte enrichment. Apical in follicle cell epithelia | Oocyte enrichment | ✓ |
| osk | Oocyte enrichment (Ephrussi et al., 1991) | ND | ✓ |
NA: not analyzed; ND: no data available; NE: no expression; nc: nurse cells; ✓: these genes show localization in ovaries or embryos (Table 1) compatible with their being transported by the BicD/Egl machinery. Known Egl targets are grk, nos, insc, osk..
Validation of top Egl target candidates
We validated the 28 top candidates in independent experiments by qPCR (Fig. 1E), comparing their enrichment again to known BicD/Egl targets as well as to non-localizing transcripts from the same IPs. 27 of these 28 putative novel Egl target candidates turned out to be enriched in anti Egl::GFP IPs compared to the mock IPs. The fold enrichment values from the qPCR analysis cannot be compared directly with the ones from the sequencing experiment because they are normalized and calculated using different parameters (see methods). Nevertheless, both experimental methods revealed a higher enrichment score than the known Egl targets for most of the top hits. The enrichment was also observed when comparing the Egl::GFP IPs to the total input mRNA (not shown). Only the enrichment of RpS29 could not be confirmed. The fact that this mRNA showed also no specific localization pattern by in situ hybridization (see bellow, Tables 1 and 2) suggests that this is a false positive hit. This indicates that our approach to find novel targets gives consistent results even with different analysis tools and that results are reproducible in independent IPs.
Egl target mRNAs are enriched for specific localization patterns
BicD/Egl is needed to transport mRNAs from the nurse cells into the oocyte and to the apical region in embryos. To validate the data set we therefore analyzed the subcellular localization of the top 50 mRNAs in ovaries and embryos. We searched for known embryonic and ovarian localization patterns in the literature and the following databases: Fly-Fish6 (http://fly-fish.ccbr.utoronto.ca), BDPG (http://insitu.fruitfly.org/cgi-bin/ex/insitu.pl) and DOT11 (http://tomancak-srv1.mpi-cbg.de/DOT/main.html). The different localization patterns found for Egl candidates are summarized in Table 1 and 2. snmRNA:331/snRNA:7SK and snoRNA:U3:9B were not analyzed and the expression of CG34357 was bellow detection. Because pbl expression was only studied in embryos, the localization of 47 top candidates was studied in embryos and 46 top candidates in ovaries.
Because the purification was performed with embryonic extracts as starting material, we expected the identified mRNAs to show specific localization patterns during embryogenesis. 10 mRNAs localized to the embryonic pole plasm and/or pole cells, 10 localized apically of the syncytial blastoderm nuclei and 4 of them showed both localization patterns (Table 1, examples in Figs. 2, 3). Interestingly, these locations are cellular compartments where components of the BicD/Egl complex can also be found enriched.10 Surprisingly however, Egl protein showed a strong posterior localization (Fig. 2A), while posterior anti BicD staining was only slightly elevated (Fig. 2B) and sometimes even not recognizably.12 Examples of mRNAs showing preferential localization to the pole plasm and pole cells are shown in Fig. 2C.
Figure 2.
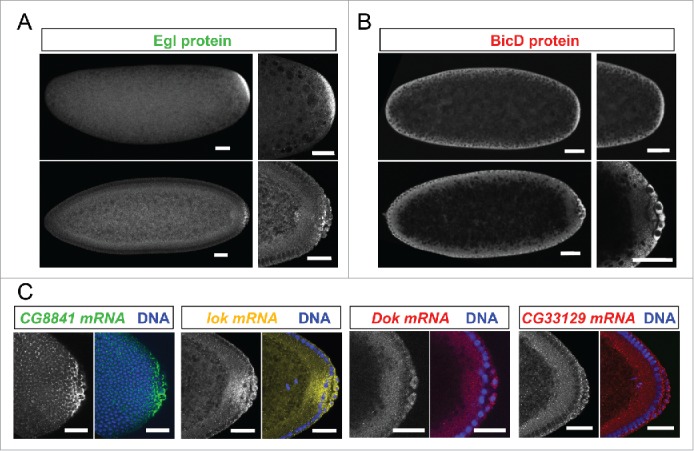
Egl targets are enriched for mRNAs localizing to pole plasm and pole cells. 21% (10/47) of the BicD/Egl targets localize to the pole plasm or pole cells compared to 8.4% of a random set.6 (A) Egl protein is enriched in the pole plasm (upper panels, embryo in syncytial division before polar bud formation) and in pole cells (lower panels, embryo in late stage 4). (B) BicD protein signal is detected slightly enriched in pole plasm and pole cells. Stages are as in A). (C) In situ hybridization to whole-mount wild-type embryos (wt, OreR) using antisense RNA probes for the candidate mRNAs CG8841, lok, Dok and CG33129. Hoechst (blue) visualizes the DNA. Scale bars are 30 μm.
Figure 3.
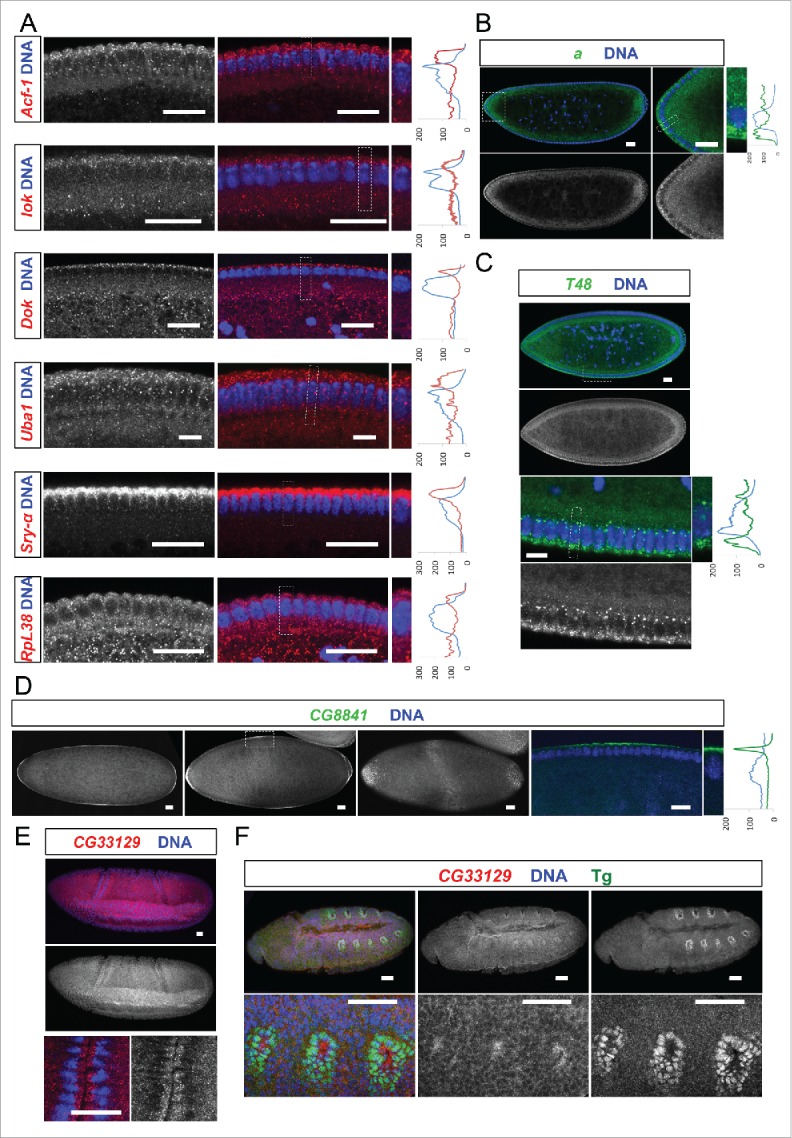
Egl targets show specific and novel apical localization patterns. Apically localizing mRNAs are 6-fold enriched compared to a published random set (10/47; 21% vs 3.4%6). In situ hybridizations to wild-type (OreR) embryos oriented anterior to the left and dorsal up. Hoechst (blue) visualizes the DNA. (A) Novel transcripts showing continuous apical localization in blastoderm embryos (red signal). Apical localization varies and changes for different mRNAs and nuclear cycles, respectively. RpL38 mRNA in situ was used as a control that shows the distribution of a non-localizing mRNA. (B-D) Novel candidates expressed apically but not continuously from anterior to posterior. (B) arc (a) is enriched apically of the nuclear layer in the anterior region of the embryo (green signal). (C) T48 is enriched apically only in the ventral region (green signal). (D) CG8841 shows apical enrichments at the anterior and posterior poles (including pole cell expression) and in the middle of the embryo (green signal). CG33129 (red signal) shows apical localization in the invaginating furrows during gastrulation (E) and in tracheal precursor cells (stained for the Tango (Tg) marker in green) later in embryogenesis (F). Scale bars represent 25 µm except in (F) were they represent 30 µm. Histograms depicted on the right side of the pictures show the fluorescence intensity along the apical-basal axis of the marked region around a blastoderm nucleus (dotted square). This region is also shown magnified on the right side.
Among the apically localizing mRNAs some showed a continuous apical distribution along the a-p body axis. These are Acf1, lok (chk2/mnk), Dok, Sry-a and Uba1 (Fig. 3A). Others, however, were enriched apically but displayed a non-uniform pattern of expression along one of the body axes (Fig. 3B–D). arc (a) mRNA was enriched apically in the anterior region of the embryo (Fig. 3B) and consistent with this localization, Arc protein was also localized apically in several epithelia.13 Arc has been reported to be a component of adherens junctions and to be required for eye and wing development.13 T48 mRNA signal was enriched apically, but only in the ventral region. This apical localization remarkably correlates with its protein localization and its function in ventral furrow formation (Fig. 3C;14). CG8841 is apically enriched at the anterior and posterior poles, in a dorsal-ventral stripe in the middle of the embryo, and it accumulates in the pole cells (Fig. 3D, Fig. 2C). Interestingly, the CG33129 mRNA shows apical localization in invaginating furrows during gastrulation and also in tracheal precursor cells later in embryogenesis (Fig. 3E–F). The apical enrichment of these mRNAs in cells that are undergoing morphogenetic movements suggests that localization of this mRNA and the protein it encodes might play a role in the apical movement of these cells.
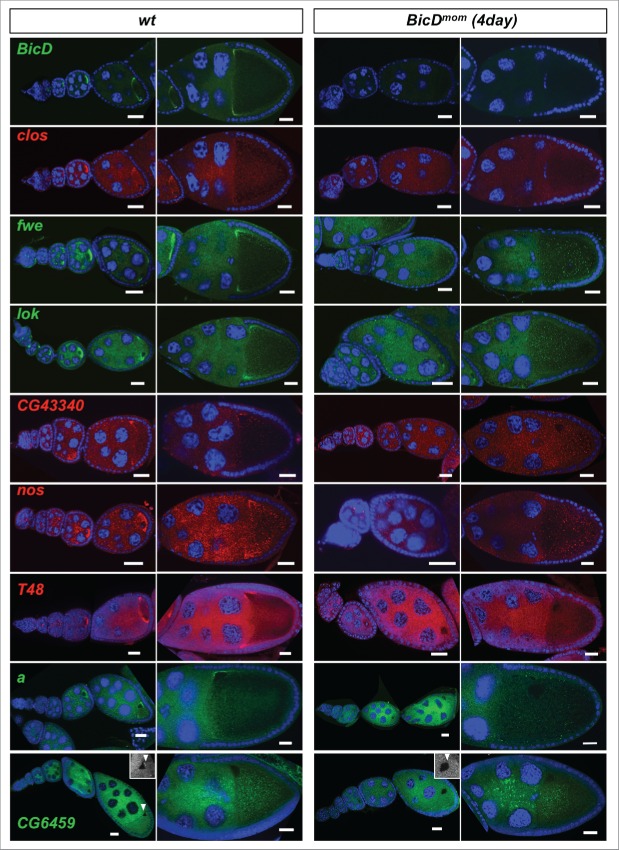
in situ hybridization experiments revealed that 28 of the 46 (61%) mRNAs expressed in ovaries showed clear enrichment in the oocyte relative to the nurse cells at some point during oogenesis (Table 2, Figs. 4, 5 and Fig. S2). Eight additional mRNAs (17%) displayed strong oocyte staining even though the oocyte signal was not higher than the nurse cell signal (Table 2, Fig. 4 and Fig. S2). Because transcription in the female germ line is almost exclusively happening in nurse cell nuclei, this distribution pattern most likely requires active transport as well. Consistently, we also found that 79% of the Egl::GFP target candidates were expressed maternally in embryos (Table 1). Interestingly, many oocyte enriched mRNAs displayed specific subcellular localization patterns. Most of them localized posteriorly up to stage 6 and then relocalized to the anterior cortex (Table 2, Figs. 4, 5 and Fig. S2). Additionally, 3 of the oocyte-enriched candidates were also expressed in the somatic follicle cells of the ovary where they localized apically (Table 2, Fig. 5A). Work with different mRNAs had shown previously that this localization pattern is also dependent on dynein.15 In summary, the present knowledge about the function of BicD/Egl in mRNA localization can explain for about 80% of the top candidates (Table 2) why they have been isolated.
Figure 4.
Egl targets are enriched for mRNAs that localize to the oocyte in a BicD dependent manner. 61% of the BicD/Egl targets tested showed accumulation in the oocyte compared to the expected 17% observed in a random set of mRNAs. In situ hybridization to wild-type (OreR) controls and to egg chambers 4 d after turning off BicD expression (BicDmom). Antisense RNA probes for the candidate mRNAs were labeled with green or red fluorescent signals. Almost no BicD mRNA signal is observed when BicD expression is off (green signal, upper-most panels). During early wild-type oogenesis, clos, fwe, lok, CG43340, nos, T48, and a accumulated in the oocyte where they became enriched at the posterior till stage 6. Subsequently the signal appeared at the anterior cortex by stage 7, showing this pattern until mid to late oogenesis (left panels). These mRNAs failed to efficiently accumulate in BicDmom oocytes when BicD was off and the late localization in the oocyte was also severely impaired in BicDmom oocytes (right panels). CG6459 showed expression in the oocyte cytoplasm at all stages. However, in this case the oocyte signal was not enriched compared to its accumulation in nurse cells. A weak BicD dependent concentration of the mRNA at the dorsal side of the oocyte nucleus was observed by stage 8 (arrowhead, and magnified region in inset picture). By stage 9 a more clear presence of a dotted CG6459 signal was observed in the oocyte, but levels did not seem to exceed nurse cell levels. In BicDmom ovarioles big blobs of CG6459 mRNA signal were seen in the nurse cell cytoplasm, suggesting a problem in transport of this mRNA. Hoechst (blue) visualizes the DNA. Scale bars are 20 µm.
Figure 5.
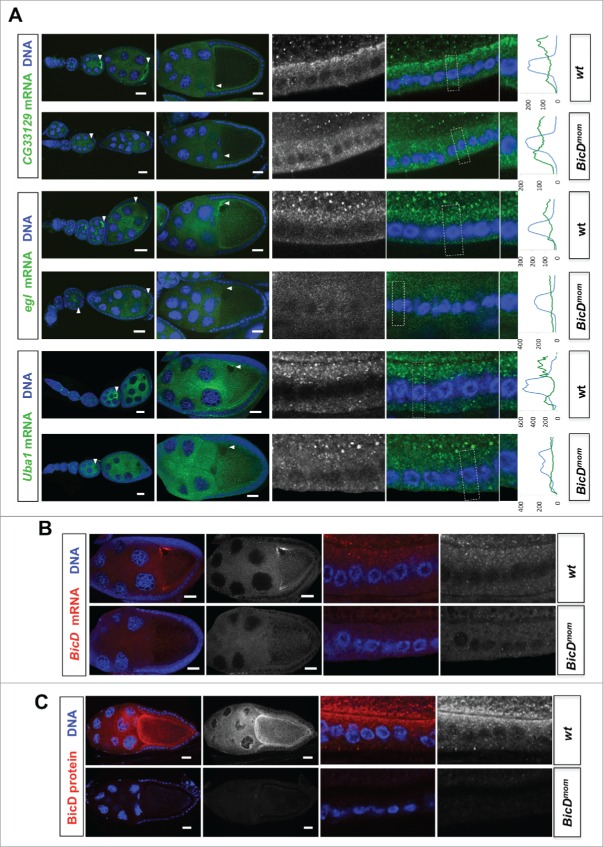
BicD is needed for apical mRNA localization in follicle cells. 6.5% of the mRNA tested show apical localization in follicle cells. This mRNAs class is enriched 8-fold among the Egl::GFP targets compared to a random set (6.5% vs 0.8%11) (A) In situ hybridization to wild-type (OreR, wt) controls and to egg chambers 4 d after turning off BicD expression (BicDmom). Oocyte enrichment (arrows) as well as apical follicle cell localization of CG33129, egl and Uba1 was impaired when BicD was off. High magnification of the stage 10 follicle cell epithelium is shown in the right panels. Note that apical is toward the oocyte (up in the magnified pictures). Scale bars are 20 µm. Uba1 apical enrichment is weaker and less cortical, but still requires BicD. Histograms depicted on the right side of the pictures show the fluorescence intensity along the apical-basal axis of the marked cortical region around the nuclear layer (dotted square). This region is also shown magnified on the right side of the micrographs. (B) BicD mRNA expression (red) is drastically reduced in stage 9–10 BicDmom egg chambers in the germline and also in the somatic follicle cells (high power pictures on the right). Note that BicD mRNA is also apically localized as expected since it is also a target of Egl. (C) BicD protein (red) is also apically enriched in wild-type follicle cells, but is almost undetectable in BicDmom egg chambers, oocytes and follicle cells. Hoechst (blue) visualizes the DNA. Scale bars are 20 µm.
To systematically analyze whether the BicD/Egl targets are enriched for specific gene ontology (GO) groups we also compiled an extended top target list. Sorted according to cellular components, the members of this list were significantly (p < 0.05) enriched for mRNAs that encode cytoplasmic proteins, components of the pole plasm and of ribonucleoprotein granules (Fig. S3A). Among the biological processes, developmental processes such as germ cell development, oocyte and embryonic axis specification, cell fate determination and cell maturation were predominant (Fig. S3B). Interestingly, these processes are well known for their dependence on mRNA localization. The enrichment of molecular functions is, however, less clear (Fig. S3C). While nucleotide and RNA binding proteins might be expected, we presently do not know why calmodulin binding proteins made it to the top of the list.
Localization of Egl target candidates requires BicD
To test whether the localization of the candidate mRNAs indeed depends on the BicD/Egl localization machinery we used the BicDmom flies described previously to turn off BicD expression in the germ line once the oocyte is made.16,17 Around 4 d after shutting down BicD expression, ovaries contained egg chambers with strongly reduced or undetectable levels of BicD mRNA and BicD protein (Figs. 4, 5B–C and17). Indeed, at the time BicD mRNA became undetectable, oocyte enrichment of the candidate mRNAs was severely impaired (Fig. 4), indicating that the top candidate Egl::GFP targets indeed depend on BicD for their localization. mRNAs from egl, CG33129 and Uba1 showed oocyte enrichment as well as apical localization in the somatic follicle cells. Interestingly, their localization was affected in both cell types in BicDmom ovaries (Fig. 5A). Because BicD mRNA (Figs. 4, 5B) and BicD protein (Fig. 5C) levels were not only reduced in the germ line, but also in the follicle cells, it appears that the BicD/Egl machinery also localizes mRNAs apically in the somatic tissue of the female gonad.
To study the involvement of BicD in localizing mRNAs that are enriched in the embryonic pole plasm, we studied transcript localization in embryos from mothers carrying a dominant BicDIIIE48 allele. Embryos laid by such mothers lack anterior structures and many of them show a strong double abdomen phenotype called bicaudal.18 A large proportion of the BicD protein gets mislocalized to the anterior of these embryos, producing ectopic anterior localization of the posterior mRNA determinants osk and nos, 2 BicD/Egl targets.12,19 lok mRNA is enriched in the pole plasm and pole cells of wild-type embryos20 (Fig. 2C, Fig. S4). Embryos from BicDIIIE48/r5 mothers showed additional enrichment of lok mRNA at the anterior of the young embryo (Fig. S4) providing good evidence that the lok mRNA is a target of BicD/Egl in embryos as well.
Putative localization elements in Egl target mRNAs
mRNA localization motives facilitating the binding to the BicD/Egl localization machinery could so far not be predicted based on their primary sequence. NMR studies of the K10 TLS localization signal showed that it forms a stem-loop with 2 double-stranded RNA helices adopting an unusual A′-form conformation.21 These helices were associated with runs of 3 or more purines on one side of the stem. Consistently, stem-loops within the mapped localization elements of well known BicD/Egl targets, such as grk, ftz, h, bcd, wg, osk and the I-factor RNAs, contain 2 or more stretches of at least 3 contiguous purines on the same side of the stem.21,22 Based on this we set up a bioinformatics approach to predict functionally important helices of this kind in our candidate mRNA data set. We expect such signals to be conserved between different Drosophila species. Accordingly, orthologous sequences from 10 Drosophila species were aligned based on their secondary structure prediction. Conserved stem-loops were then sorted for the presence of 2 or more stretches of at least 3 contiguous purines on the same side of the stem. Using this approach we optimized the program by searching for the known BicD/Egl dependent localization hairpins (Fig. S5). For each hairpin we then plotted the conservation across Drosophila species against the percentage of bases that make pair bonds for each hairpin (this serves as a simplified measurement of the hairpin stability). We found that in the case of h, orb, grk, K10, osk and bcd the best predicted localizing A'-form hairpin (best conserved and with highest frequency of base matches) coincided with the known hairpins responsible for apical or oocyte localization of the transcripts(21,22; Fig. S5). In the case of wg and ftz the program still predicted the known localization sequences and they were among the best scoring ones, but not at the top. It is possible that in these cases the presence of redundant localization signals or the high conservation intrinsic to open reading frames (ORFs) was distorting the values (see Discussion). We also consider it possible that the 2 best predicted hairpins in ftz and wg are the main localization elements in several other Drosophila species, but have become partially redundant with new ones in melanogaster.
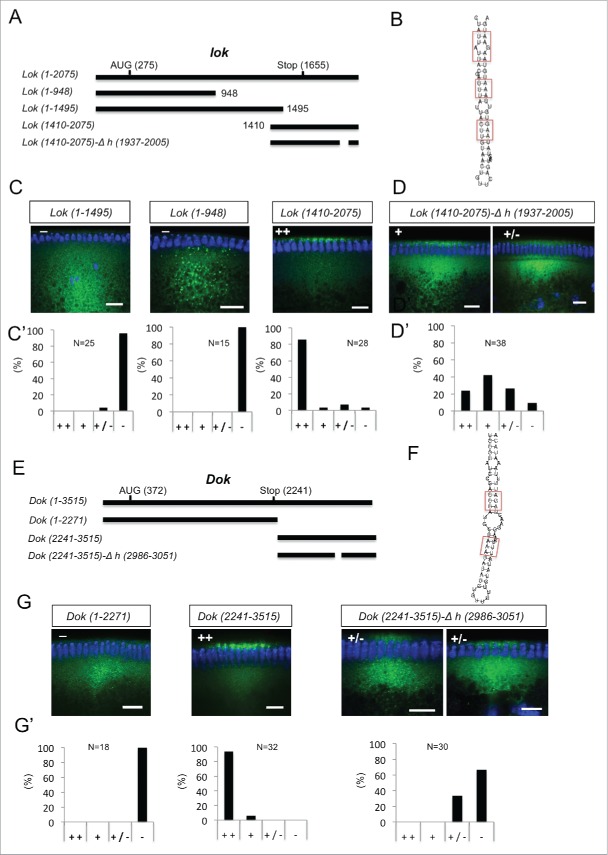
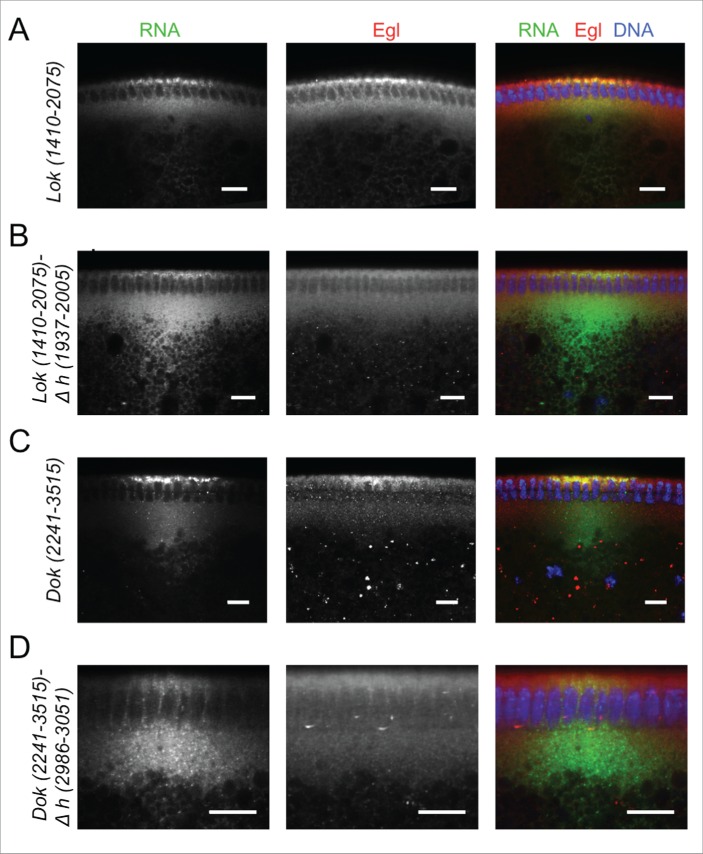
We next analyzed the Egl:GFP targets that showed a clear oocyte, pole plasm or apical enrichment in ovaries or embryos for the presence of conserved predicted A'-form helices (Fig. S6 and Fig. S7). Two mRNAs, pgc and sry-α, showed the presence of only one putative A'-form type helix, and this was identified in their ORF (Fig. S7). lok, Dok, fs(N)1 and T48 have one most likely localization hairpin that shows the best folding energy and is highly conserved (Fig. S6A) The analysis of CG33129, egl, a, mus210/Xpc, insc, dap and fatp/CG7400 revealed that although the best-scoring hairpin might likely be the localizing signal, some other hairpins would probably need to be tested as well (Fig. S6 and Fig. S7). Similarly CG43340, didum, CG12488, Uba1, Acf1, CG8841, fwe and hts have at least 2 putative A'-form helices with high scores (Fig. S6 and Fig. S7). We selected Dok and lok to test the localization activity of the predicted hairpins (Fig. 6) by injecting fluorescently labeled RNAs into the basal cytoplasm of syncytial embryos and following their apical transport. For lok, the most highly conserved A'-form hairpin resides in the ORF, but there is also one in the 3′UTR that, although less conserved, has the highest frequency of base matches (Fig. S6A). Only lok sequences containing the 3′UTR supported apical localization, while fragments containing the ORF regions did not localize (Fig. 6A, C C',). Furthermore, deletion of the predicted conserved A'-form in the 3′ UTR greatly impaired apical localization of the 3′UTR region (Fig. 6B, D, D'). Similarly, the best predicted A'-form hairpin of Dok was also identified in the ORF (Fig. S6A), but the ORF RNA alone did not support apical transport (Fig. 6E, G, G'). In contrast, the 3′UTR localized strongly to the apical side (Fig. 6E, G, G') and the deletion of its predicted 3′UTR A'-form stem loop abolished most of the transport (Fig. 6E–G, G'). Egl protein is recruited to its target RNAs injected into the basal region of the embryo and it gets co-transported apically with the RNA.10 Injecting lok and Dok 3′ UTRs, but not the corresponding constructs lacking the localizing hairpins, also led to an apical enrichment of Egl above the site of injection (Fig. 7A–D). The bioinformatics predictions can therefore reveal functional localization elements in the 3′ UTR that serve to localize these mRNAs apically by the BicD/Egl transport machinery.
Figure 6.
3′ UTR hairpins of Dok and lok are responsible for apical localization. (A, E) Schematic representations of the test lok (A) and Dok (E) mRNAs injected. (B, F) Secondary structure of the localizing hairpins in lok (B) and Dok (F) predicted using the RNAfold web server. (C, D, G) Representative images of embryos injected with the respective lok (C-D) and Dok (G) constructs indicated above each picture. Fluorescently labeled transcripts (green signal) were injected into blastoderm embryos basally and fixed 12 min thereafter. Images are oriented with apical at the top. Hoechst (blue) visualizes the DNA. The categorization of the RNA enrichment to the apical cytoplasm for the corresponding injections shown in (C, D, G) is shown under each embryo image (C', D', G'). The percentage of embryos showing strong (++), weak (+), very weak (+/−) or no apical (−) localization is shown for each construct. (N) Number of embryos scored for each mRNA injection. Scale bars are 10 µm.
Figure 7.
Recruitment of Egl to apically localizing Dok and lok 3′ UTRs. (A-D) Immunostaining revealed the distribution of Egl (red) following basal injection of lok (green) (A, B) and Dok RNA fragments (C, D) into wild-type embryos. Egl is co-recruited to apically localized Dok and lok 3′ UTR sequences (A, C), but not if the localizing hairpin sequence was deleted from the construct (B, D). Scale bars are 10 µm.
Discussion
We have identified BicD/Egl mRNA targets in 0–8 hour old Drosophila embryos and found that 80% of the top candidates showed localization patterns that are compatible with their being actively transported during oogenesis and/or during the first 4 hours of embryogenesis by the BicD/Egl dependent localization machinery. At present we do not know for most of the remaining transcripts whether they are transported by Egl during different developmental stages, whether a competing process prevents their accumulation at the expected target site or whether the Egl-mediated transport also leads to yet uncharacterized distribution patterns. For CG33129, however, we observed that the transcripts are also localized apically in later stages in the tracheal precursors and insc is known to be localized in older embryos apically in the neuroblasts.3,4 Compared to a random group of mRNAs,6 the BicD/Egl top targets described here show a 6-fold enrichment for apically localizing embryonic mRNAs (10/47; 21% vs 3.4%). Similarly 21% (10/47) of the BicD/Egl targets are also enriched for mRNAs that localize to the pole plasm or pole cells compared to the reported 8.4% of a random set.6 Compared to a random set of mRNAs11 we also observed a 3–4-fold increase in oocyte accumulation among the BicD/Egl targets (61% vs 17%) and an 8-fold increase in apical follicle cell localization (6.5% vs 0.8%) although our sample size for this phenotype is very small (n = 3). The same localization patterns were also enriched in a statistical analysis that took into account a much larger group of Egl targets (see Fig. S3D, E). In contrast, basally localized transcripts were clearly underrepresented in our data set (Fig. S3D). The enrichment for specific mRNA localization patterns in ovaries and embryos indicates that the BicD/Egl machinery specifically acts for nurse cell to oocyte transport, for apical transport of mRNAs during oogenesis and embryogenesis, and for enrichment in the pole plasm and pole cells. All these processes indeed involve a polarized MT network and the BicD/Egl/Dynein machinery. These results further imply that different subcellular localization patterns identified for other mRNAs are generated by different mechanisms or at least require an additional element. The multitude of different mRNA localization patterns described for instance by the high throughput screens6,11 are therefore likely to involve additional mechanisms such as mRNA diffusion coupled with entrapment and degradation, and active transport utilizing different transport systems.
The data set of apically localized mRNAs bound to Egl can be grouped into 3 functional groups: 1) Genes with nuclear functions, involved in DNA metabolism, DNA repair, chromosome structure and DNA damage response. 2) Genes coding for proteins that associate directly or indirectly with the plasma membrane. 3) Genes coding for secreted proteins. The first group includes the genes like lok that encodes the Drosophila Chk2 kinase involved in monitoring DNA damage, Mutagen-sensitive 210 (Mus-210, also Xpc), which encodes a protein with DNA binding domains and a predicted DNA repair function, and Acf1 (ATP-dependent chromatin assembly factor 1), which is involved in the assembly and maintenance of heterochromatin.20,23,24 Also belonging to this group of nuclear proteins are the transcription factors encoded by the segmentation genes h and ftz. At the time the proteins encoded by these mRNAs become active, their target nuclei are also positioned apically. It thus appears that the localization of their mRNAs facilitates an efficient reaching of their target site by the encoded protein. Furthermore, as for the apically localized mRNAs with striped localization patterns like ftz and h (but also wg, CG8841, a, T48), the apical localization of these transcripts might serve to avoid lateral diffusion of their products to the neighboring nuclei and compartments. For example, Acf1 is a subunit of the nucleosome-remodeling complex involved in chromatin assembly and chromatin-mediated gene repression, and in Acf1 mutants heterochromatin assembly is disturbed.23 Interestingly, when pericentric heterochromatin forms during the blastoderm stages, it is localized to the apical side of the nuclei, adjacent to where we found Acf1 mRNA localized. Because Acf1 mRNA localization depends on BicD activity,25 it appears that BicD and egl might also play a role in heterochromatin assembly by localizing Acf1 mRNAs in embryos.
The second group contains mRNAs encoding proteins that associate with the plasma membrane. The PDZ domain protein Arc is a component of the adherens junctions. Protein and mRNA reside apically in several epithelia, and in embryos they are found in regions that undergo morphogenetic movements, such us invagination, elongation or convergent extension.13 Serendipity α (Sry-α) is associated with the plasma membrane and with actin invaginations during cellularization at the blastoderm stage.26 Dok is a signaling protein required upstream of JNK signaling during dorsal closure. It is found at the cell cortex associating with the plasma membrane where it is ideally positioned to respond to Src signaling.27 T48 is a transmembrane protein that is needed together with G protein signaling to recruit adherens junctions and cytoskeletal regulators to sites of apical constriction during ventral furrow formation.14 Remarkably, CG8841, the novel pole plasm component that displays additional apical mRNA localization, encodes a predicted transmembrane domain protein, too.
The localization of most Egl target mRNAs in ovaries depends on BicD. This testifies to the quality of the data presented in the candidate list and it suggests that aside from the top candidates, the list may contain numerous additional targets with lower scores. Indeed, known BicD/Egl targets, like Chc, orb, K10, h and ftz appear further down in the list with lower enrichment scores (Table S1). While validating the BicD/Egl targets we noticed that BicD is also required for apical localization of many mRNAs in “normal” somatic cells (aside from the syncytial embryo). However, loss-of-function mutations in egl and BicD were initially isolated as female sterile mutants because of their essential function in the female germ line.28 The fact that we now also found Egl target mRNAs that are apically localized in developing tracheal cells and in salivary glands (data not shown), together with the known role for BicD in localizing insc mRNA in neuroblasts, suggest that this machinery is also working in somatic embryonic and larval cells and tissues where a polarized cytoskeleton is present. This indicates that mRNA localization by BicD/Egl is at work in many different cell types, but that it is more important in large cells, such as the female germ line, the syncytial embryo and the nervous system. This is consistent with the BicDnull phenotype, which reveals important zygotic functions for this gene. Most BicDnull animals die during larval stages but a very small fraction makes it to adulthood. Such adults are uncoordinated and lethargic and die within 2 d.29
Taking advantage of the new pool of verified Egl target mRNAs and of the published NMR structure of the K10 localization signal, we also set up a bioinformatics approach to identify A'-form hairpin structures in Egl-bound mRNAs. Interestingly, although most of the conserved A'–form hairpins were predicted to be located in the ORF of the mRNAs (Fig. S6D), we could not validate their functionality in the 2 cases we tested. Instead A'–form hairpins identified in the 3′UTR turned out to serve as functional localization elements. This suggests that giving priority to the hairpins found in the 3′ UTR should improve the chance of finding the correct localization sequences. One reason for this could be that hairpins of ORFs that are in a state of repressed translation - bound to translational repressors or to a repressed translation machinery - may be masked such that the transport machinery cannot bind to them. In contrast, the 3′ UTR may be free of stalled ribosomes and accessible for recruitment by the transport complex. Another possibility is that additional sequences in the 3′UTR serve as binding site for additional factors that may facilitate Egl binding. Similarly, embedding the localization signal in the context of the 3′UTR, which is more AT rich than the ORF, may facilitate the folding of the Egl recognition structure. This view is also supported for example by the finding that the efficient localization of h and ftz is context dependent with sequences surrounding the minimal localization signal working non autonomously, but enhancing the activity of the localization element.30,31
Material and methods
Purification of Egl bound mRNAs
RNA immunoprecipitations were performed essentially as described with some modifications.32 Briefly, protein G Sepharose beads (Gamma bind Plus agarose, Roche) were washed 3 times with blocking buffer (20 mM HEPES pH 7.9, 150 mM KCl, 20% glycerol containing 0.5% Tween-20, 1mg/ml BSA, 2 mg/ml heparin and EDTA free protease inhibitors, Roche). Beads were then blocked for 3 h at RT with the same buffer. 1 ml of monoclonal anti-GFP antibody supernatant was added per 40 μl of blocked beads and incubated for 2 h at RT with gentle rotation. Coated antibody beads were then washed with non-hypotonic buffer (20 mM HEPES pH 7.9, 2mM MgCl2, 150 mM KCL, 1mM DTT, 20% glycerol containing 0.5% Tween and EDTA free protease inhibitors, Roche). For extract preparation, 1 gr of 0–8 h old embryos were dechorionated and homogenized in 2 ml hypotonic buffer (20 mM HEPES pH 7.9, 2 mM MgCl2, 10 mM KCL, 1 mM DTT containing 0.5% Tween-20, EDTA free protease inhibitors, Roche, and RNase inhibitor, Biolabs, 100 units/ml). All further steps were performed at 4°C. The homogenized extract was cleared at 10,000 × g for 20 min. For each IP 323 μl of salt adjusting buffer (20mM HEPES pH 7.9, 57% glycerol, 0.4M KCl, 2mM MgCl2, 1mM DTT) and 2 μl of RNase free DNase I (20 U/ml, Roche) were added to 600 μl of cleared supernatant. This salt adjusted extract was added to 40 μl of coupled antibody beads and incubated overnight at 4°C with rotation. The immunoprecipitate was washed 8 times with high salt wash buffer (20 mM HEPES pH 7.9, 2 mM MgCl2, 200 mM KCL, 20% Glycerol, 1mM DTT containing 0.5% Tween, EDTA free protease inhibitors and RNase inhibitor (Biolabs, 100 units/ml)), rotating it for 10 min, followed by centrifugation at 2,000 rpm for 2 min. Beads were treated with 30 μg of Proteinase K (Roche) in proteinase K buffer (150 mM NaCl, 12.5 mM EDTA, 10 % SDS, 0.1M Tris-HCL pH 7.5) in a total volume of 100 μl for 30 min at 55°C. RNA was extracted using the Trizol reagent (Invitrogen) following the manufacture's instructions. The integrity and quality of the co-immunoprecipitated RNA was assessed using the Agilent RNA 6000 Nano Kit on an Agilent 2100 bioanalyzer (Agilent Technologies) following the manufacturer's instructions.
Preparation of the library for sequencing
The material of 2 independent IPs was pooled and used for library preparation (15 µl of RNA with 47,7 ng/µl for the control IP and with 73,7 ng/µl for the Egl::GFP IP) following the manufacture's instructions (TruSeq RNA Sample Prep Kit V2, Illumina, La Jolla, USA).
RT-PCR and RT-qPCR analysis
The RNA from each IP was resuspended in 25 μl of DEPC treated water and its quality was analyzed with the Eukaryote Total RNA Nano chip in an Agilent bioanalyzer. RT-PCR assays were performed to optimize the RNA IP protocol. For this, sequences were amplified using the Access RT-PCR system (Promega) and primers that spanned introns. Optimal cycle numbers were determined for each gene.15 ng of IPed RNAs were used as template for each RT-PCR reactions. Using the same amount for the Egl::GFP IPs and the control IPs allows us to normalize the total number of reads from the subsequent sequencing reaction even though it will produce an underestimated enrichment value. The enrichment for localizing mRNAs but not for housekeeping genes was also observed when using the same amount of embryonic material for normalization. No amplification was observed in reactions without RT, indicating that we do not observe amplification from DNA.
For RT-qPCR assays, cDNAs from the different samples were prepared with the SuperScript II Reverse Transcriptase and oligo dT primers (Invitrogen) following the manual's instructions and using 133 ng of IPed mRNAs. Primers used for the RT-qPCR analysis were designed in a way that one of a pair spanned an exon-junction sequence and that the amplicon length ranged from 100 to 150 bp. Sequences were amplified using the QuantiTect SYBR Green PCR Kit (Qiagen) and a Rotor Gene instrument (Qiagen). Calculation of mRNA levels was done using the 2 [-ΔΔC(T)] method.33,34 Fold enrichment (mRNA levels present in the IP over mRNAs levels in the mock IP) was plotted. C(T) values used were the means of duplicate technical and biological repeats.
Bioinformatics analysis of Illumina data
The two single-end RNAseq libraries produced 49 M and 52 M reads with a read length of 100bp on an Illumina HiSeq2000 Instrument (Illumina, La Jolla, USA). The quality of the reads was assessed using FastQC (version 0.10.1, www.bioinformatics.babraham.ac.uk/projects/fastqc) and reads mapping to rRNA genes (∼80%) were removed. The remaining reads (9,5 M and 10,8 M reads) were mapped to the fly genome (version 5.45 of the Flybase) using the spliced alignment approach implemented in TopHat2 version 2.0.5.35 For each annotated gene (annotation version 5.45 of Flybase) we counted the number of reads mapping to it using the program HTSeq-count (www.w-huber.embl.de/users/anders/HTSeq, version 0.5.3). To test for significant differences of gene expression levels between the Egl::GFP and the y w control the R÷package DESeq was used.36 More precisely, raw read counts were normalized to the total number of reads, and the variation of read counts per gene was assumed to follow the variation of counts across genes. The reported p-value is corrected for multiple testing following Benjamini and Hochberg.37 Fold enrichment represent the ratio between the normalized reads from the Egl IP over the control IP. GO term enrichment analysis was done with the website http://geneontology.org.
Fluorescent in situ hybridization (FISH) to whole mount embryos and ovaries and immunostainings
Plasmids (BDGP resources;38) containing the cDNA of the candidate genes were linearized with the corresponding restriction enzymes (New England Biolabs) and used as templates to generate digoxigenin- or Fluorescein (FITC)-labeled RNA antisense probes. In situ hybridization experiments on ovaries and embryos were performed essentially as described39,40 but 5% milk powder (Rapilat) in PBT (PBS, 0.1% Tween) was used as blocking reagent. The digoxigenin labeled probes were detected with sheep-anti-digoxigenin antibodies and Cy3-conjugated donkey anti-sheep IgG F(ab')2 fragments (Jackson Immunoresearch). Fluorescein-labeled probes were detected with mouse anti-fluorescein antibodies (Roche) and Cy3-conjugated goat anti-mouse antibodies (Jackson Immunoresearch). For double RNA in situs, Cy3-conjugated anti-mouse antibodies and Cy5-conjugated anti-sheep antibodies were used (Jackson Immunoresearch). Immunostainings were done using the following primary antibodies: mouse anti-BicD (1B11, 1: 10 dilution;41) anti-Egl (1:5,000 dilution;42). Where required, nuclei were stained for 20 min with 2.5 µg/ml of Hoechst 33258 during the final wash step. Images were analyzed with a Leica TCS-SP8 or Leica TCS-SP5 confocal microscopes. Different pictures were taken for each egg chamber in an ovariole focusing on the oocyte center. The picture showing the entire ovariole was then assembled in Adobe Photoshop. All patterns described were observed reproducibly and were observed in most of the embryos and ovaries analyzed. Histograms depicting the fluorescence along the apical-basal axis were done using Fiji software.
Fluorescent RNA synthesis and injections
RNAs for injections were synthesized in the presence of Alexa-488-UTP (Molecular probes) as described.10 Dok and lok RNAs were synthesized using LD32155 and LD27875 plasmids, respectively (BDGP resources). The Dok (1–2271) DNA template was generated by linearizing the corresponding plasmid with BamHI. The lok plasmid was linearized with SphI or BamHI and used as template to generate lok (1–948) and lok (1–1495) transcripts. Deletion of the corresponding localizing hairpins in Dok and lok 3′ UTR was done on LD32155 and LD27875 by site directed mutagenesis. Wild-type and deleted forms of the 3′ UTR regions were amplified by PCR and used as templates for transcription. Fluorescently labeled RNAs were used at 0.5–1 μg μl−1. Fixing was done 12 min after injection of the last embryo and immunofluorescent staining of injected embryos was done as described.10 Images were analyzed with a Leica TCS-SP8 confocal microscope. Strong localization means that most of the signal was in the apical cytoplasm. Weak localization means most signal remained basally, but there were some apical caps of signal; “in very weak localization” apical cap signals were barely above background fluorescence of the basal cytoplasm while no fluorescent puncta were observed in the apical cytoplasm in “no apical localization.”
Stem loop predictions
The sequences of the candidate genes were retrieved from FlyBase (release 5.1). The longest isoform of every gene was chosen. Local secondary structures were explored by means of RNALfold, from the Vienna package43 (version 2.1.8), with the following options: 25°C temperature and no “lonely pairs.” A conservation score for the Drosophila melanogaster genome was available on the UCSC genome browser.44 It was computed by means of PhastCons45 on the multiple alignment of the dm3 genome with:- D. simulans (droSim1)- D. sechellia (droSec1)- D. yakuba (droYak2)- D. erecta (droEre2)- D. ananassae (droAna3)- D. pseudoobscura (dp4) - D. persimilis (droPer1)- D. willistoni (droWil1)- D. virilis (droVir3) - D. mojavensis (droMoj3) - D. grimshawi (droGri2)- A. gambiae (anoGam1) - A. mellifera (apiMel3) - T. castaneum (triCas2)
Predicted hairpins were further analyzed for the presence of at least 2 A/G stretches, each at least 3 nucleotides in length. Overlapping hairpins were removed, retaining the ones with the lowest energy. Hairpins with a conservation score higher than 0.5 and longer than 40 nt were reported.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank S. Bullock for the Egl::GFP flies and for teaching us how to do RNA injections in embryos. We also thank R. Lehmann for the anti-Egl antibodies and the Drosophila Genomics Resource Center (supported by NIH grant 2P40OD010949-10A1) for fly stocks.
Funding
This work was supported by the Swiss National Science Foundation [grant number 31003A_153280]; the Canton of Bern to B.S; the “Mittelbauvereinigung” of the University of Bern (MVUB); and an Equal Opportunity grant from the Phil.-nat. Fakultät to P.V.P.
References
- 1.Vazquez-Pianzola P, Suter B. Conservation of the RNA transport machineries and their coupling to translation control across eukaryotes. Comparative Functional Genomics 2012; 2012:287852-13; PMID:22666086; http://dx.doi.org/ 10.1155/2012/287852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weil TT. mRNA localization in the Drosophila germline. RNA Biol 2014; 11:1010-8; PMID:25482896; http://dx.doi.org/ 10.4161/rna.36097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes JR, Bullock SL, Ish-Horowicz D. Inscuteable mRNA localization is dynein-dependent and regulates apicobasal polarity and spindle length in Drosophila neuroblasts. Curr Biol 2004; 14:1950-6; PMID:15530398; http://dx.doi.org/ 10.1016/j.cub.2004.10.022 [DOI] [PubMed] [Google Scholar]
- 4.Simmonds AJ, dosSantos G, Livne-Bar I, Krause HM. Apical localization of wingless transcripts is required for wingless signaling. Cell 2001; 105:197-207; PMID:11336670; http://dx.doi.org/ 10.1016/S0092-8674(01)00311-7 [DOI] [PubMed] [Google Scholar]
- 5.Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 2001; 105:209-19; PMID:11336671; http://dx.doi.org/ 10.1016/S0092-8674(01)00312-9 [DOI] [PubMed] [Google Scholar]
- 6.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007; 131:174-87; PMID:17923096; http://dx.doi.org/ 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 7.Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 2009; 23:1546-58; PMID:19515976; http://dx.doi.org/ 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suter B, Romberg LM, Steward R. Bicaudal-D, a Drosophila gene involved in developmental asymmetry: localized transcript accumulation in ovaries and sequence similarity to myosin heavy chain tail domains. Genes Dev 1989; 3:1957-68; PMID:2576013; http://dx.doi.org/ 10.1101/gad.3.12a.1957 [DOI] [PubMed] [Google Scholar]
- 9.Stuurman N, Häner M, Sasse B, Hübner W, Suter B, Aebi U. Interactions between coiled-coil proteins: Drosophila lamin Dmo binds to the Bicaudal-D protein. Eur J Cell Biol 1999; 78:278-87; PMID:10350216; http://dx.doi.org/ 10.1016/S0171-9335(99)80061-2 [DOI] [PubMed] [Google Scholar]
- 10.Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 2001; 414:611-6; PMID:11740552; http://dx.doi.org/ 10.1038/414611a [DOI] [PubMed] [Google Scholar]
- 11.Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife 2015; 4:R106; PMID:25838129; http://dx.doi.org/2590944 10.7554/eLife.05003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wharton RP, Struhl G. Structure of the Drosophila BicaudalD protein and its role in localizing the posterior determinant nanos. Cell 1989; 59:881-92; PMID:2590944; http://dx.doi.org/ 10.1016/0092-8674(89)90611-9 [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Lengyel JA. Drosophila arc encodes a novel adherens junction-associated PDZ domain protein required for wing and eye development. Dev Biol 2000; 221:419-34; PMID:10790336; http://dx.doi.org/ 10.1006/dbio.2000.9689 [DOI] [PubMed] [Google Scholar]
- 14.Kölsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science 2007; 315:384-6; PMID:17234948; http://dx.doi.org/ 10.1126/science.1134833 [DOI] [PubMed] [Google Scholar]
- 15.Horne-Badovinac S, Bilder D. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genetics 2008; 4:e8; PMID:18208331; http://dx.doi.org/ 10.1371/journal.pgen.0040008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swan A, Suter B. Role of Bicaudal-D in patterning the Drosophila egg chamber in mid-oogenesis. Development 1996; 122:3577-86; PMID:8951073; http://dx.doi.org/ 10.1242/dev.099432 [DOI] [PubMed] [Google Scholar]
- 17.Vazquez-Pianzola P, Adam J, Haldemann D, Hain D, Urlaub H, Suter B. Clathrin heavy chain plays multiple roles in polarizing the Drosophila oocyte downstream of Bic-D. Development 2014; 141:1915-26; PMID:24718986; http://dx.doi.org/ 10.1242/dev.099432 [DOI] [PubMed] [Google Scholar]
- 18.Mohler J, Wieschaus EF. Dominant maternal-effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics 1986; 112:803-22; PMID:3082713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ephrussi A, Dickinson LK, Lehmann R. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 1991; 66:37-50; PMID:2070417; http://dx.doi.org/ 10.1016/0092-8674(91)90137-N [DOI] [PubMed] [Google Scholar]
- 20.Oishi I, Sugiyama S, Otani H, Yamamura H, Nishida Y, Minami Y. A novel Drosophila nuclear protein serine/threonine kinase expressed in the germline during its establishment. Mechanisms Dev 1998; 71:49-63; PMID:9507063; http://dx.doi.org/ 10.1016/S0925-4773(97)00200-1 [DOI] [PubMed] [Google Scholar]
- 21.Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A'-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Structural Mol Biol 2010; 17:703-9; PMID:20473315; http://dx.doi.org/ 10.1038/nsmb.1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jambor H, Mueller S, Bullock SL, Ephrussi A. A stem-loop structure directs oskar mRNA to microtubule minus ends. RNA 2014; 20:429-39; PMID:24572808; http://dx.doi.org/ 10.1261/rna.041566.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chioda M, Vengadasalam S, Kremmer E, Eberharter A, Becker PB. Developmental role for ACF1-containing nucleosome remodellers in chromatin organisation. Development 2010; 137:3513-22; PMID:20843858; http://dx.doi.org/ 10.1242/dev.048405 [DOI] [PubMed] [Google Scholar]
- 24.Masrouha N, Yang L, Hijal S, Larochelle S, Suter B. The Drosophila chk2 gene loki is essential for embryonic DNA double-strand-break checkpoints induced in S phase or G2. Genetics 2003; 163:973-82; PMID:12663536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Börner K, Jain D, Vazquez-Pianzola P, Vengadasalam S, Steffen N, Fyodorov DV, Tomancak P, Konev A, Suter B, Becker PB. A role for tuned levels of nucleosome remodeler subunit ACF1 during Drosophila oogenesis. Dev Biol 2016; 411:217-30; PMID:26851213; http://dx.doi.org/ 10.1016/j.ydbio.2016.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweisguth F, Lepesant JA, Vincent A. The serendipity α gene encodes a membrane-associated protein required for the cellularization of the Drosophila embryo. Genes Dev 1990; 4:922-31; PMID:2166703; http://dx.doi.org/ 10.1101/gad.4.6.922 [DOI] [PubMed] [Google Scholar]
- 27.Biswas R, Stein D, Stanley ER. Drosophila Dok is required for embryonic dorsal closure. Development 2006; 133:217-27; PMID:16339186; http://dx.doi.org/ 10.1242/dev.02198 [DOI] [PubMed] [Google Scholar]
- 28.Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster: II Mutations blocking oogenesis or altering egg morphology. Genetics 1991; 129:1119-36; PMID:1783295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ran B, Bopp R, Suter B. Null alleles reveal novel requirement for Bic-D during Drosophila oogenesis and zygotic development. Development 1994; 120:1233-42; PMID:8026332 [DOI] [PubMed] [Google Scholar]
- 30.Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J 2003; 22:2484-94; PMID:12743042; http://dx.doi.org/ 10.1093/emboj/cdg230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snee MJ, Arn EA, Bullock SL, Macdonald PM. Recognition of the bcd mRNA localization signal in Drosophila embryos and ovaries. Mol Cell Biol 2005; 25:1501-10; PMID:15684399; http://dx.doi.org/ 10.1128/MCB.25.4.1501-1510.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA 2007; 13:1198-204; PMID:17592038; http://dx.doi.org/ 10.1261/rna.563707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; http://dx.doi.org/ 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/23618408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 35.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14:R36; PMID:23618408; http://dx.doi.org/ 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11:R106; PMID:20979621; http://dx.doi.org/ 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Royal Statistical Society Series B-Methodological 1995; 57:289-300 [Google Scholar]
- 38.Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al.. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res 2007; 17:1823-36; PMID:17989253; http://dx.doi.org/ 10.1101/gr.6679507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes SC, Krause HM. Single and double FISH protocols for Drosophila. Methods Mol Biol 1999; 122:93-101; PMID:10231786 [DOI] [PubMed] [Google Scholar]
- 40.Vazquez-Pianzola P, Urlaub H, Suter B. Pabp binds to the osk 3′UTR and specifically contributes to osk mRNA stability and oocyte accumulation. Dev Biol 2011; 357:404-18; PMID:21782810; http://dx.doi.org/ 10.1016/j.ydbio.2011.07.009 [DOI] [PubMed] [Google Scholar]
- 41.Suter B, Steward R. Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell 1991; 67:917-26; PMID:1959135; http://dx.doi.org/ 10.1016/0092-8674(91)90365-6 [DOI] [PubMed] [Google Scholar]
- 42.Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev 1997; 11:423-35; PMID:9042857; http://dx.doi.org/ 10.1101/gad.11.4.423 [DOI] [PubMed] [Google Scholar]
- 43.Lorenz R, Bernhart S, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler P, Hofacker I. ViennaRNA Package 2.0. Algorithms Mol Biol 2011; 6:26; PMID:22115189; http://dx.doi.org/ 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al.. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 2014; 42:D764-70; PMID:24270787; http://dx.doi.org/ 10.1093/nar/gkt1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005; 15:1034-50; PMID:16024819; http://dx.doi.org/ 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.