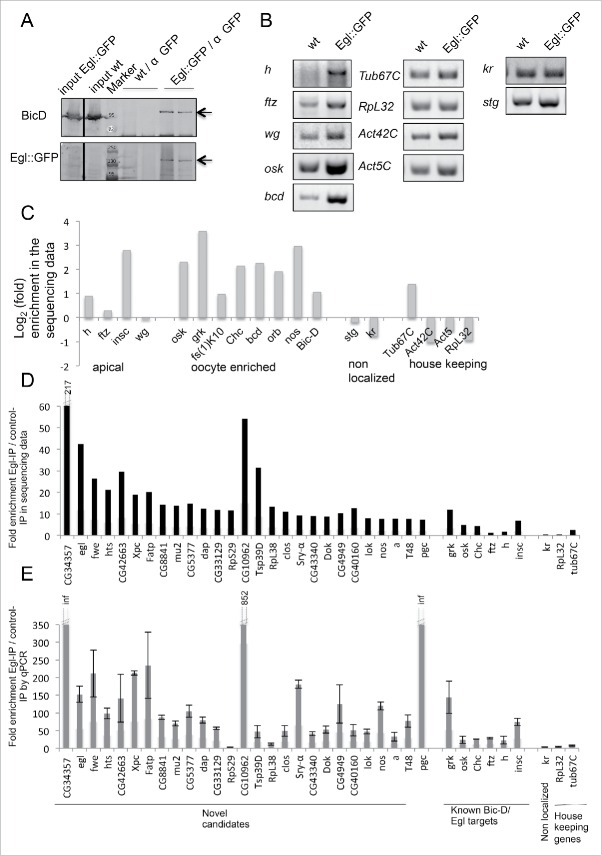
Figure 1.
RipSeq protocol retains BicD/Egl complex stability and enriches for known target mRNAs. (A-B) Extracts from Egl::GFP expressing embryos and wt control extracts were subjected to IP with anti-GFP antibodies. Western blotting tested for the presence of the BicD and Egl::GFP (A). Semiquantitative RT-PCR tested for the enrichment of known BicD/Egl mRNA targets and for the lack of enrichment of non-localizing mRNAs and house keeping mRNAs (B). (C) Rip-Seq enrichment of mRNAs from the test set. (D-E) Most top candidate BicD/Egl targets identified by Rip-Seq (D) could be validated by RT-qPCR analysis in an independent IP experiment (E). Error bars represent +/− SD of 2 independent IPs. Note that fold enrichment values of the 2 different experiments cannot be compared directly because they are calculated using different formulas (see methods). Nevertheless, enrichments in the Egl::GFP IPs relative to mock IPs is reproducible.