Figure 1.
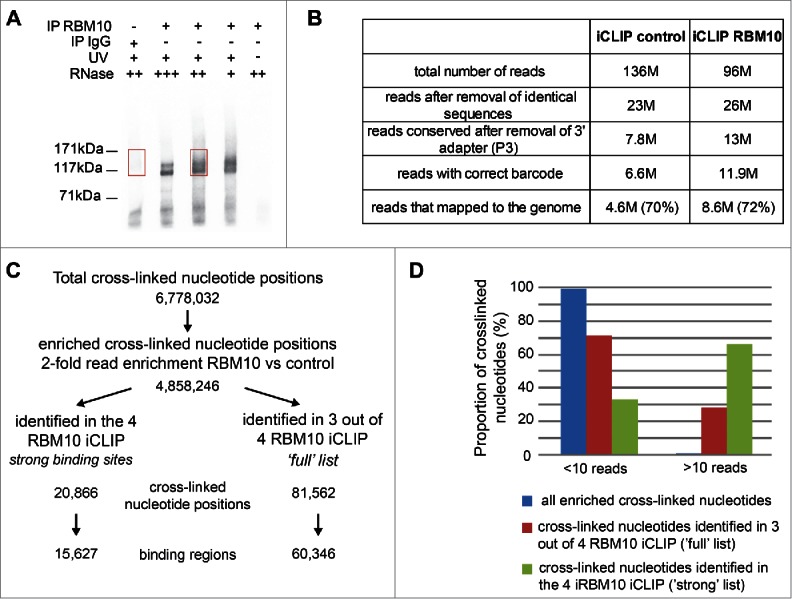
Genome-wide mapping of RBM10 RNA-binding sites in a mouse mandibular MEPA cell line using iCLIP. (A) Autoradiograph of cross-linked protein/RNA complexes after immunoprecipitation and 32P RNA labeling. Immunoprecipitations were performed with an antibody that recognizes both isoforms of RBM10 protein or with IgG, as a negative control. Three different dilutions of RNase I were used. Cells untreated with UV were also used as a control. Sections of the membrane cut for the library preparation are indicated by red boxes (B) Number of reads obtained for each iCLIP experiment after processing of the sequencing data (removal of PCR duplicates, trimming of adapter sequences, attribution of the read to one specific replicate using the barcode) (C) Number of cross-linked nucleotides obtained through all iCLIP experiments and the ones reproducibly found in several replicates of the iCLIP experiments. (D) Graph showing the proportion of cross-linked nucleotides supported by more than 10 reads in the list of binding sites present in all 4 iCLIP experiments (‘strong’ list) or in 3 out of 4 experiments (‘full’ list).
