Figure 4.
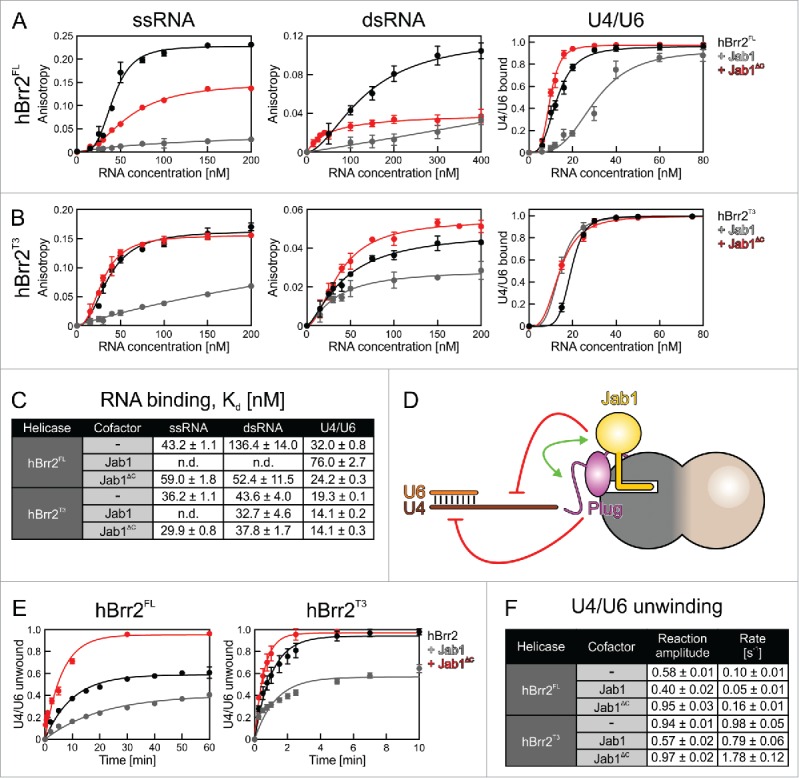
Effects of the NTR and the Prp8 Jab1 domain on RNA binding and unwinding by Brr2. (A,B) Assays monitoring RNA binding by hBrr2FL (A) or hBrr2T3 (B). Black curves – hBrr2FL or hBrr2T3 alone; gray curves – hBrr2FL or hBrr2T3 in complex with Jab1; red curves – hBrr2FL or hBrr2T3 in complex with JabΔC. Left panels – binding to ssRNA; central panels – binding to dsRNA; right panels – binding to yU4/U6 snRNA. Binding of ssRNA and dsRNA was monitored by FP using 5-FAM-labeled RNAs. Binding of yU4/U6 was monitored via EMSA. Data points and error bars represent means +/− SEMs of at least 2 independent experiments. (C) RNA affinities. Values represent means +/− SEMs of at least 2 independent experiments. Kd values were obtained by fitting the data from (A) and (B) to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient).50 (D) Scheme summarizing the effects of the NTR and the Jab1 domain on different portions of a substrate. Red symbols – inhibition; green double-arrow – reinforcement. (E) U4/U6 unwinding by Brr2 variants in the absence or presence of Jab1 variants. Black curves – Brr2FL or Brr2T3 alone; gray curves – Brr2FL or Brr2T3 in complex with Jab1; red curves – Brr2FL or Brr2T3 in complex with JabΔC. (F) Unwinding parameters. Amplitudes and unwinding rates were obtained by fitting the data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). Values represent means +/− SEMs of at least 4 independent experiments.
