Figure 5.
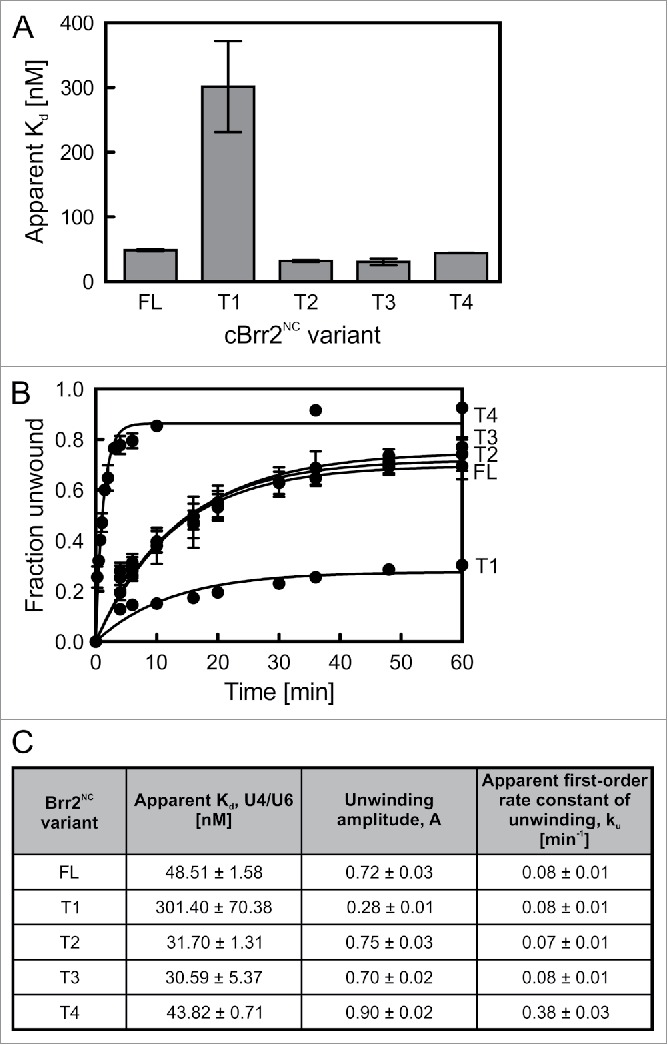
Effects of NTR truncations on C. thermophilum Brr2NC variants. (A) Binding of N-terminal Brr2NC truncations to U4/U6 di-snRNA monitored by electrophoretic mobility shift assays. Values represent means ± SEMs of at least 2 independent experiments. Kd values were obtained by fitting quantified binding data to a single exponential Hill function (fraction bound = A[protein]n/([protein]n+Kdn); A – fitted maximum of RNA bound; n – Hill coefficient). (B) Time courses of U4/U6 unwinding by Brr2NC variants. Data represent means ± SEM of at least 3 independent experiments. Amplitudes and unwinding rates were obtained by fitting the quantified data to a first-order reaction (fraction unwound = A[1-exp{-ku t}]; A – amplitude of the reaction; ku – apparent first-order rate constant of unwinding; t – time). (C) U4/U6 di-snRNA affinities and unwinding parameters of Brr2NC variants. Values represent means ± SEM of at least 2 independent experiments.
