Figure 3. mRNA structure downstream of SS promotes efficient -1 PRF in the copA gene.
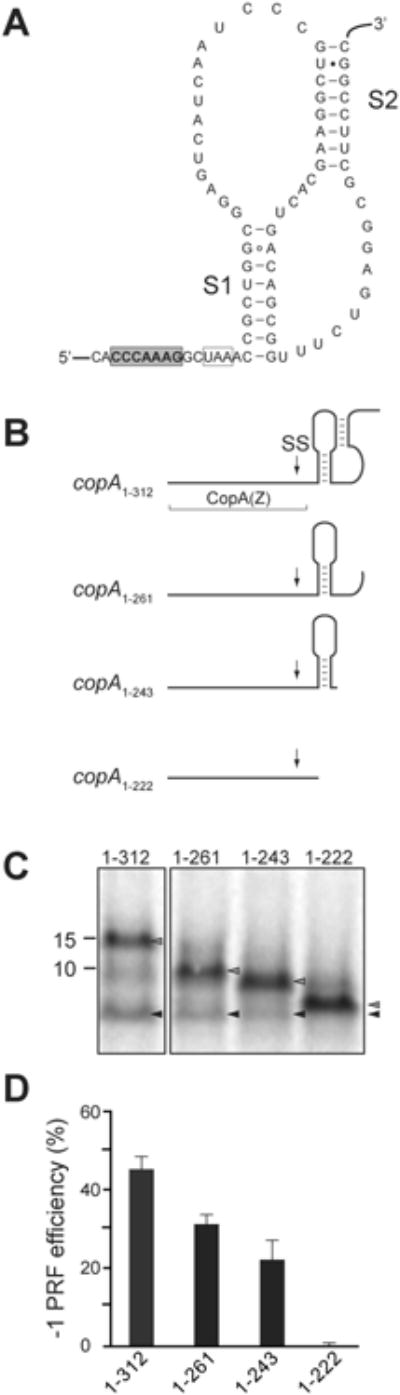
(A) The predicted structure of the pseudoknot in the copA mRNA downstream from the SS. The slippery site is shaded in gray, the -1 frame stop codon is boxed, and stems S1 and S2 of the pseudoknot are indicated.
(B) Predicted secondary structure of the mRNA segments downstream of the SS in the 3′-truncated copA templates. The boundaries of the CopA(Z) coding segment is indicated and the location of SS is marked by an arrow.
(C) SDS-gel analysis of the [35S]-labeled protein products expressed in vitro from the 3′-truncated copA templates shown in (B). Grey arrowheads indicate the bands corresponding to the full-sized 0 frame product; black arrowheads indicate CopA(Z) generated via -1 PRF.
(D) Quantification of the -1 PRF efficiency in the different 3′-truncated copA templates from the relative intensity of the bands indicated in (C). Error bars show standard deviation from the mean in 3 independent experiments.
See also Figure S3.
