Figure 4. The CopA nascent peptide contributes to the high efficiency of -1 PRF.
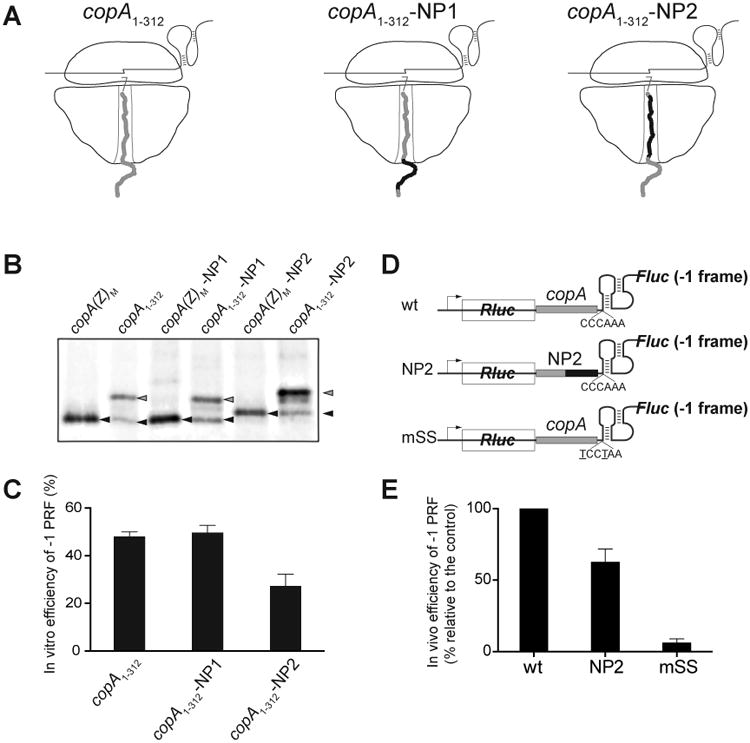
(A) Cartoon representation of the ribosomes translating through the copA SS sequence of the wt template (copA1-312) or of the NP1 and NP2 mutants. The sequence of the wt CopA nascent chain is shown in grey and the mutant peptide sequence is shown in black. The nucleotide and amino acid sequences of the templates are show in detail Figure S1.
(B) Gel electrophoretic analysis of the [35S]-labeled truncated CopA encoded in the 0 frame of the copA1-312 wt or mutant templates (gray arrowheads) and CopA(Z) generated via -1 PRF (black arrowheads). The reference templates (M) encode the marker CopA(Z) proteins whose sequences matched those generated from the mutant templates via -1 PRF. The sequences of the reference templates are shown in Figure S1.
(C) Quantification of the -1 PRF efficiency in the different copA templates from the relative intensity of the bands shown in (B). Error bars show deviation from the mean in 3 independent experiments.
(D) The structures of the dual luciferase reporters containing unaltered (wt) or mutant (NP2) CopA segments. The construct carrying the copA sequence with the mutated SS (mSS) was used as a negative control.
(E) Efficiency of the in vivo -1 PRF in the dual luciferase reporters shown in (D). Error bars show standard deviation from the mean in 3 independent experiments.
See also Figure S1.
