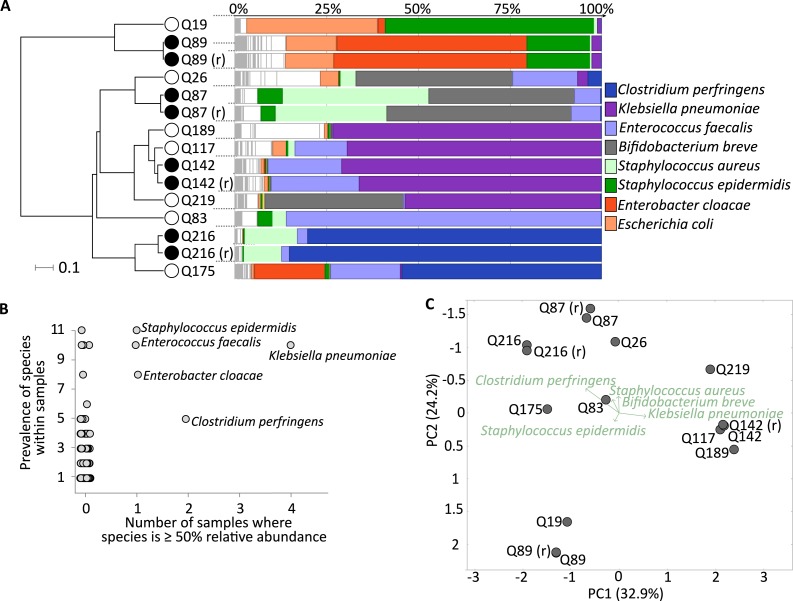
Figure 1. Healthy premature infant gut microbiome.
(A) Metagenomic profiles for the eleven preterm samples and four replicates at the species level. Samples clustered by UPGMA using Bray–Curtis distances shown on left, with replicates highlighted by filled nodes. Relative abundances by rank order shown on right, with the top 8 most abundant species coloured and labelled, leaving remaining species in white. (B) Dominant species, based on ≥50% abundance, shown on x-axis, with overall prevalence of the species across samples shown on y-axis. Sample number reflects eleven neonates as replicates are averaged. The five labelled species are present in five or more samples with at least one in >50% abundance. (C) Principal coordinates analysis (PCoA) using Bray–Curtis distances at the species level for all fifteen samples. Separation of the three broad sample groups shown by biplot of the top five species.