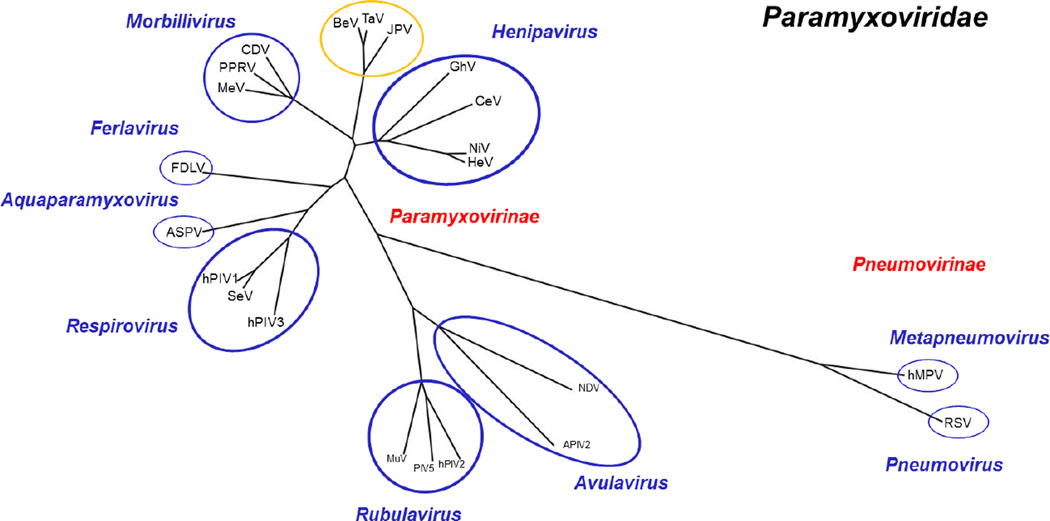
Fig. 1. Paramyxoviridae polymerase protein phylogeny tree.
Polymerase/Large protein sequences of selected viruses were acquired from the NCBI Protein Database and Virus Pathogen Resource (ViPR). L protein sequences were aligned using the COBALT Multiple Alignment Tool-NCBI. Aligned sequences were then used to generate a phylogenic tree using COBALT NCBI TreeView1.8. The generated tree was visualized and modified using the FigTree Program. Abbreviations: APIV-2, avian parainfluenza virus 2; ASPV, Atlantic salmon paramyxovirus; BeV, Beilong virus; CDV, canine distemper virus; CeV, Cedar Virus; FDLV, Fer-de-Lance virus; GhV, Ghana virus; HeV, Hendra virus; hMPV, Human metapneumovirus; hPIV1, Human parainfluenza virus 1; hPIV2, Human parainfluenza virus 2; hPIV3, Human parainfluenza virus 3; JPV, J Paramyxovirus; PIV5, Parainfluenza virus 5; RSV, respiratory syncytial virus; MeV, Measles virus; MuV, Mumps virus; NDV, Newcastle disease virus; NiV, Nipah Virus; PPRV, Peste-des-petits ruminants virus; TaV, Tailam Virus. Paramyxoviridae family (black); sub-families (red); genus (blue); unclassified (yellow).