Abstract
The TOR (target of rapamycin) proteins play important roles in nutrient signaling in eukaryotic cells. Rapamycin treatment induces a state reminiscent of the nutrient starvation response, often resulting in growth inhibition. Using a chemical genetic modifier screen, we identified two classes of small molecules, small-molecule inhibitors of rapamycin (SMIRs) and small-molecule enhancers of rapamycin (SMERs), that suppress and augment, respectively, rapamycin's effect in the yeast Saccharomyces cerevisiae. Probing proteome chips with biotinylated SMIRs revealed putative intracellular target proteins, including Tep1p, a homolog of the mammalian PTEN (phosphatase and tensin homologue deleted on chromosome 10) tumor suppressor, and Ybr077cp (Nir1p), a protein of previously unknown function that we show to be a component of the TOR signaling network. Both SMIR target proteins are associated with PI(3,4)P2, suggesting a mechanism of regulation of the TOR pathway involving phosphatidylinositides. Our results illustrate the combined use of chemical genetics and proteomics in biological discovery and map a path for creating useful therapeutics for treating human diseases involving the TOR pathway, such as diabetes and cancer.
Keywords: drug discovery, drug target identification, proteomics, diabetes
The development of experimental methods to modulate selectively the properties of proteins is central to defining the principles that underlie signaling networks (1–9). Although phenotype-based chemical genetic screens have been highly successful in identifying conditional probes for dynamic cellular processes (10) as well as potential leads to therapeutic drugs (11), subsequent target identification is currently the rate-limiting step in this approach. Using the budding yeast Saccharomyces cerevisiae as a model system, we illustrate herein an efficient small-molecule target identification strategy for chemical genetics that relies on proteome chips. We apply this strategy to the discovery of putative intracellular targets of small molecules that modify the cellular effects of rapamycin, a Streptomyces hygroscopicus polyketide macrolide that is a promising anti-cancer drug (12).
TOR (target of rapamycin) proteins are phylogenetically conserved from yeast to humans, and are members of the phosphatidylinositol kinase (PIK)-related kinase family (13), which includes the DNA-damage checkpoint proteins ATM (mutated in ataxia telangiectasia), ATR (ATM-related), and DNA-PKcs [mutated in severe combined immunodeficiency (SCID)]. TOR is a central regulator of cell growth in response to nutrient signals (14). The TOR-dependent nutrient-response network controls many aspects of metabolism, the deregulation of which may lead to diseased states. For example, organ transplant patients treated with rapamycin have been found to develop hyperglycemia and hyperlipemia (www.wyeth.com), a state characteristic of non-insulin-dependent diabetes mellitus. We developed a high-throughput phenotype-based screen to search for chemical genetic modifiers of the rapamycin-sensitive functions of TOR. By identifying small molecules that selectively modify the rapamycin-sensitive pathways, we hope to gain a better understanding of the cellular effects of rapamycin and, ultimately, to be able to modulate TOR function in vivo. Here, we report the discovery of small-molecule inhibitors of rapamycin (SMIR) that impinge on the TOR network. To gain insight into the molecular mechanisms of these small molecules, we performed gene expression profiling and probed proteome chips with the small molecules. These studies identified genetic modifiers of the TOR-signaling network and suggest a mechanism of regulation of the TOR pathway involving phosphatidylinositides.
Materials and Methods
Chemical Genetic Screen. Wild-type (rapamycin-sensitive) yeast cells EGY48 or BY4741 were inoculated into yeast extract/peptone/dextrose/adenine (YPDA) containing 100 nM of rapamycin and robotically dispensed at 40 μl per well into clear-bottomed 384-well plates by using a Multidrop 384 (LabSystems, Chicago). We used 384-pin arrays (Genetix, Boston) to transfer 50–100 nl of compound per pin from the library plates to assay plates. Thus, the final concentration of the library compound assayed was estimated to be ≈100 μM. The assay plates were incubated at 30°C, and yeast growth was scored by visual inspection once a day for 5 days. Compounds responsible for the distinct growth phenotypes were then retested to confirm the activity.
Transcript Profiling of Yeast and Human Cells Treated with Rapamycin and SMIRs. Exponentially growing cells were treated with rapamycin for 30 min in the presence or absence of SMIRs. The final concentration of rapamycin was 100 nM for yeast and 20 nM for human Jurkat cells. RNAs were prepared according to Affymetrix protocols and used for hybridization to Ye6100 Arrays or Hu35K Arrays (Affymetrix, Santa Clara, CA).
Yeast Proteome Chip Probing. The yeast proteome chips were prepared as described (15). To block the proteome chips before probing, the chips were immersed in PBS buffer with 1% BSA at 4°C overnight without shaking. After the chips were washed three times with 50 ml of fresh PBS buffer, 180 μl of 200 μM SMIR3-biotin or 10 μM SMIR4-biotin in PBS buffer containing 1% BSA and 0.1% Nonidet P-40 was added to the proteome chips. The binding reactions were carried out in a humidity chamber at room temperature for 1 hour without shaking. The chips were then washed extensively with a large volume of PBS buffer containing 0.1% Nonidet P-40. To detect the bound small molecules on the chip, Cy3-labeled streptavidin (1:10,000 dilution) in PBS buffer was added to the surface of chips and incubated at room temperature for 30 min. The chips were then washed with a large volume of PBS buffer and spin-dried, and the image of the chips was obtained by a microarray scanner (Axon Instruments, Foster City, CA). The positives were finally identified based on the algorithm described in ref. 15.
Results and Discussion
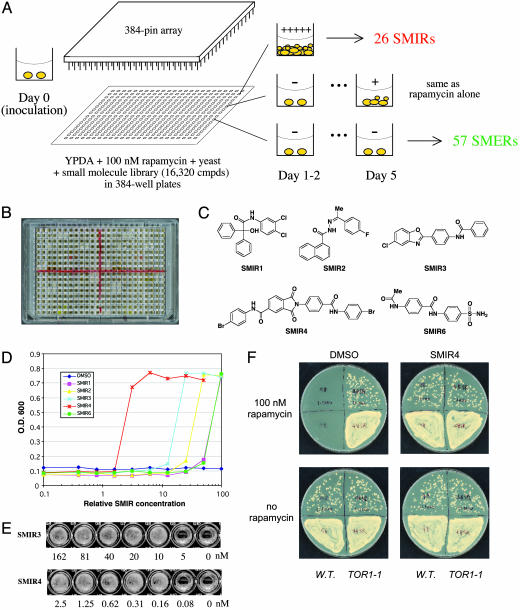
To identify small-molecule modifiers of rapamycin, we performed a chemical genetic suppressor screen (Fig. 1 A and B) using a collection of 16,320 small molecules (Diverset E, ChemBridge, San Diego). The primary screen was based on the growth inhibitory property of rapamycin in wild-type yeast cells. Small molecules were screened for their ability to rescue yeast growth in YPDA containing 100 nM rapamycin. From this screen, six compounds were identified that fully suppressed rapamycin's anti-proliferative effect (SMIRs), allowing the cells to double at a rate indistinguishable from those not treated with rapamycin. An additional 20 SMIRs (not yet characterized) were also found to restore yeast growth, although at a slower rate. The same screen also yielded 57 small molecules that caused synthetic lethality with rapamycin (SMERs) (Fig. 1A). The structures of the fast-acting SMIRs (Fig. 1C) have been confirmed by proton and carbon NMR (1H and 13C NMR), mass spectrometry, and resynthesis. Dose responses of the SMIRs are shown in Fig. 1D. Minimal inhibitory concentrations (MIC) for SMIR3 and SMIR4 (corresponding to 20 nM of rapamycin) were determined to be ≈10 nM and ≈0.16 nM, respectively (Fig. 1E).
Fig. 1.
A chemical genetic screen for small molecules that modulate rapamycin's antiproliferative effect in yeast. (A) Schematics of the screen. Compounds were transferred from library plates to assay plates (containing growth medium, rapamycin, and yeast cells) by using 384-pin arrays. SMER, small-molecule enhancers of rapamycin. (B) Retest of SMIRs in a 384-well plate. White wells indicate compound-induced yeast growth in the presence of rapamycin; black (transparent) wells indicate no growth. (C) Chemical structures of the fast-acting SMIRs (yeast growth identifiable on day 1, same as the “no rapamycin” control). (D) Dose–response curves for SMIRs in wild-type (rapamycin-sensitive EGY48) cells inoculated in YPDA containing 100 nM rapamycin. (E) Minimal concentrations of SMIR3 and SMIR4 required for yeast growth in YPDA containing 20 nM rapamycin. (F) SMIR4 treatment and the TOR1-1 (S1972R) mutation (26) both confer rapamycin resistance. Cells were plated at two different densities on the upper versus lower halves of the plates (1:1,000).
The suppressor activities of the SMIRs seem to be restricted to rapamycin, because they are unable to suppress the effects of other antiproliferatives tested, including juglone, nocodazole, and cycloheximide (data not shown). We believe that neither SMIR3 nor SMIR4 acts by altering cellular uptake or export of rapamycin, because many aspects of the rapamycin transcript profile remained (see below) in the presence of SMIR3 (at saturating concentrations) or SMIR4 (at suboptimal concentrations).
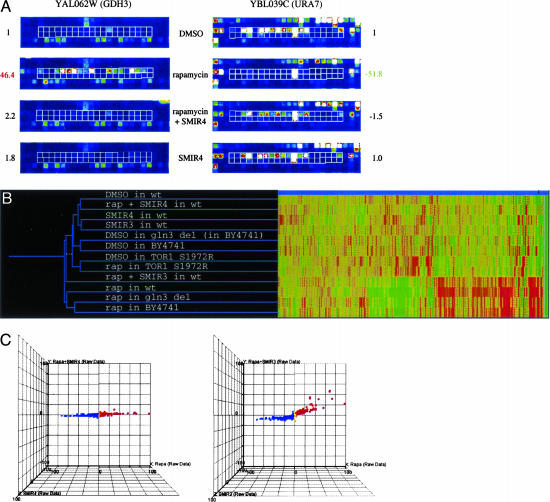
It is known that intracellular formation of a ternary complex of the immunophilin protein FKBP12, rapamycin, and the TOR protein modulates translational regulation in response to nutrient deprivation (16). More recent studies revealed that this ternary complex directly regulates a transcriptional network that responds to nitrogen and carbon sources (17–20). To elucidate the cellular pathways affected by the SMIRs, we performed genome-wide mRNA abundance-profiling experiments and compared the profiles generated from rapamycin-treated cells in the presence versus absence of the SMIRs. At a threshold of 3-fold change, 492 genes were up-regulated, and 588 genes were down-regulated upon treatment with rapamycin for 30 min (Table 1, which is published as supporting information on the PNAS web site). As shown in Fig. 2A, rapamycin induces transcription of GDH3, the NADP+-linked glutamate dehydrogenase; SMIR4 eliminates this increase in GDH3 transcription, reducing it to the level in cells not treated with rapamycin. Similarly, rapamycin-induced decrease in transcription of URA7, the CTP synthase that catalyzes the last step in pyrimidine biosynthesis, is eliminated by SMIR4. In the absence of rapamycin, SMIR4 alone has minimal effects on gene expression. In fact, nearly all changes in gene expression induced by rapamycin are completely reversed by an optimal concentration of SMIR4 (Fig. 2 B and C and Table 1). These results indicate that SMIR4 is able to reverse most cellular changes caused by rapamycin as assayed by whole-genome expression profiling. In contrast, SMIR1, -2, -3, and -6 reversed changes in the expression levels of only a subset of genes (Fig. 2B and C and Table 1). Interestingly, as we lower the concentration of SMIR4 used, its transcript profile becomes similar to that of SMIR3, suggesting that the mechanisms of action of these small molecules are related and that at high concentrations SMIR4 may have more than one target. These profiling results provide molecular evidence for an uncoupling of rapamycin-sensitive TOR functions that are essential to cell viability and those that are not. These data also suggest the possibility for using SMIRs to selectively modulate the effects of rapamycin in the cell.
Fig. 2.
SMIRs suppress rapamycin's effect in yeast on the whole-genome scale as revealed by mRNA transcript abundance analysis. (A) Example views of GeneChip features containing probes (outlined in white) specific for the GDH3 and URA7 transcripts (up- and down-regulated by rapamycin treatment, respectively) on the Affymetrix Ye6100 oligonucleotide arrays. (B) For comparing the effects of SMIRs and genetic mutations, a 2D clustergram with experiment tree and gene tree was generated in genespring (Silicon Genetics, Redwood City, CA) by using the hierarchical clustering program (minimal distance, 0.001; separation ratio, 5; new tree with standard correlation). (C) Profiling data visualized by 3D scatter plots in genespring. Fold changes over DMSO mock treatment were plotted for 6,430 genes on the Affymetrix chip. Color scaling is based on the rapamycin profile: red, increase; blue, decrease.
This ability to uncouple various aspects of TOR function may be harnessed to eliminate undesired effects of rapamycin in the clinic. In expression profiles obtained from human T cells, SMIR3 and SMIR4 were also found to suppress rapamycin-induced changes (Table 2, which is published as supporting information on the PNAS web site). These data suggest that SMIR3 and SMIR4 may target evolutionarily conserved elements between yeast and mammals.
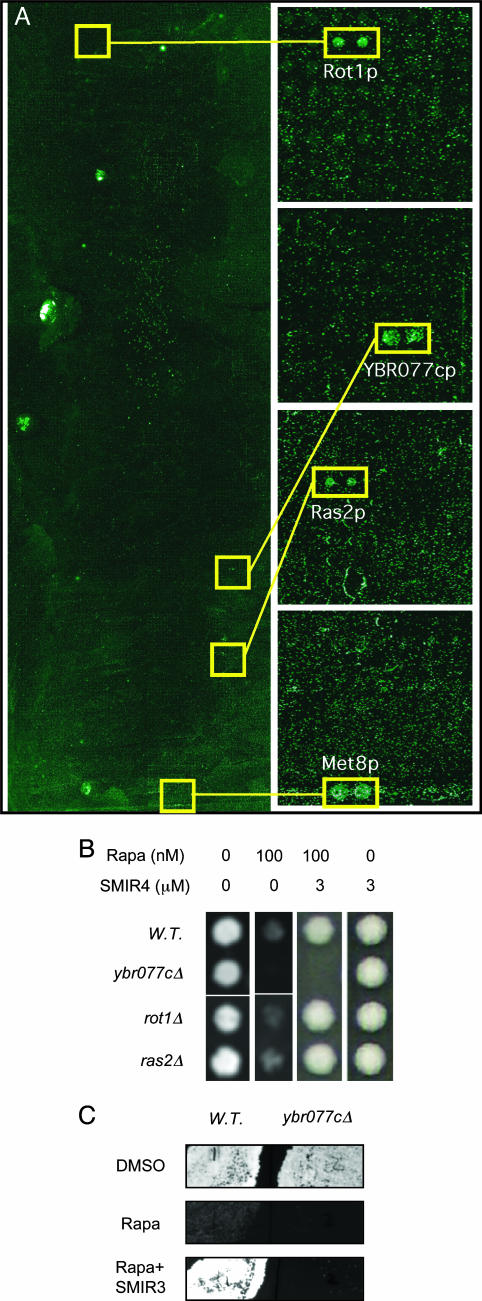
To understand the molecular mechanisms of SMIR action, we synthesized biotin-conjugated SMIR3 and SMIR4 molecules (which retained their bioactivities in vivo) and used them to probe a proteome chip containing nearly the entire yeast proteome (15) for SMIR-binding activities. Binding of biotinylated SMIR3 and SMIR4 to proteins was detected by probing with a fluorescently labeled streptavidin conjugate. Eight candidate protein targets for SMIR3 and 30 candidates for SMIR4 were identified from these experiments (Table 3, which is published as supporting information on the PNAS web site). SMIR3 binds strongly to only one protein, Tep1p, the yeast homologue of the mammalian tumor suppressor protein PTEN (phosphatase and tensin homologue deleted on chromosome 10) (21), which is a 3-phosphoinositide (PI) phosphatase (22, 23) with substrate specificities toward PI(3,4,5)P3 and PI(3,4)P2. SMIR4-binding proteins include a known suppressor of tor2 mutation (Rot1p) and 29 other proteins that were not known to be involved in Tor signaling (Fig. 3A and Table 3). Among these proteins, five are strong binders to SMIR4 based on fluorescence intensity measurements from the proteome chip.
Fig. 3.
Protein targets of SMIR4 identified by binding of SMIR4-biotin to the yeast proteome chip (15). (A)(Left) Image of the whole proteome chip. (Right) Enlarged images of the areas that contain the positive twin-spots. Four proteins that bind strongly to SMIR4 are indicated. (B and C) Yeast cell growth on agar. (B) Ybr077cp, one of the SMIR4-binding proteins identified in A, when deleted causes hypersensitivity to rapamycin and insensitivity to the suppressive effect of SMIR4, indicating that Ybr077cp is a bona fide target of SMIR4. (C) Ybr077cp is also required for SMIR3 function, suggesting possible common mechanisms by SMIR3 and SMIR4.
If one of these SMIR-binding proteins is a bona fide target of a SMIR in vivo, then elimination of that protein from yeast cells should ideally satisfy two conditions: (i) it should alter the cell's sensitivity to rapamycin, and (ii) it should reduce the ability of SMIR to suppress rapamycin's effect. To address the first condition, we tested the rapamycin sensitivity of yeast strains with deletions in each of the candidate proteins identified by the proteome chip. Of the eight SMIR3 binders, none conferred altered rapamycin sensitivity. Of the 30 SMIR4 binders, only one altered rapamycin sensitivity: Ybr077cp deletion is hypersensitive to rapamycin (Fig. 3B). YBR077C encodes a protein of unknown function and has no obvious mammalian homolog at the sequence level. As expected from the second condition for a bona fide target of SMIR4 in vivo, we found that YBR077C is required for SMIR4 to suppress rapamycin efficiently (Fig. 3B). Our result suggests that YBR077C is a new genetic modifier of the TOR pathway. Among the other SMIR4 binders, ROT1 was previously identified as a suppressor of essential cytoskeletal function only for TOR2, which is not sensitive to rapamycin (24), and thus it may not be relevant to the suppressive actions of SMIR4. The lack of a rapamycin phenotype in rot1Δ cells (Fig. 3B) is in agreement with this interpretation. Ras2p, which was one of five proteins that binds strongly to SMIR4 (Fig. 3A), was recently reported to be involved in TOR signaling (25). However, just as was observed in this work, we did not detect any change in rapamycin sensitivity in ras2Δ cells (Fig. 3B and unpublished results) and SMIR4 was fully active in ras2Δ cells (Fig. 3B). Thus, we believe that Ras2p may not be relevant to the actions of SMIR4.
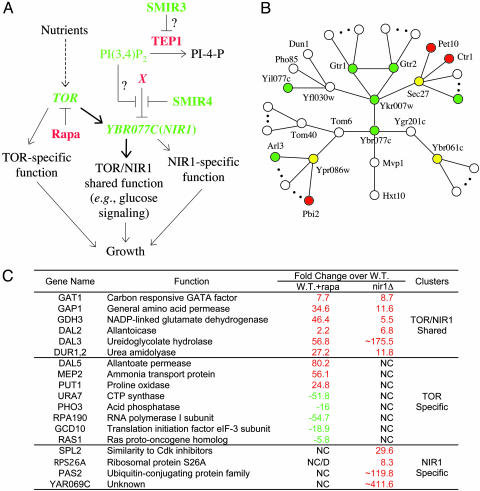
To confirm that rapamycin hypersensitivity in ybr077cΔ cells is due to inhibition of TOR, we asked whether the hypersensitivity can be suppressed by the rapamycin-resistant TOR1-1 allele (a point mutant of TOR1 that encodes a functional Tor protein that does not bind FKBP-rapamycin) (26). We transformed ybr077cΔ strain with a plasmid carrying TOR1-1 or a wild-type TOR1 control plasmid and found that TOR1-1 (but not the wild-type) allele confers complete resistance to rapamycin in ybr077cΔ cells. Also, forced expression of YBR077C rescues the hypersensitivity of ybr077cΔ cells and raises rapamycin resistance in wild-type cells (W. Zou and J.H., unpublished results). These results support YBR077C's function in the rapamycin-sensitive TOR network, which is further corroborated by the following observations. First, a transcript profile of untreated ybr077cΔ cells is strikingly similar to rapamycint-reated wild-type cells (Fig. 4C), indicating a requirement for Ybr077cp in TOR protein function. That Ybr077cp may be directly involved in TOR protein signaling is consistent with its cellular localization. We know from database (http://yeastgfp.ucsf.edu) searching that both Ybr077cp and the recently discovered Tor1p partner Kog1p (homolog of mammalian Raptor) (27) are localized to the vacuolar membrane. Next, ybr077cΔ tor1Δ double mutants exhibited multiple synthetic defects. For example, the growth rates of the double mutant spores are evidently retarded, and the double mutant cells are more sensitive to rapamycin than either single deletion mutant (data not shown), suggesting a functional interaction between YBR077C and TOR1. Lastly, we asked whether ybr077cΔ cells display any defect in TOR-dependent nutrient signaling. Indeed, we found that ybr077cΔ cells are ≈10 times less susceptible to cell death when challenged with a “glucose only” condition (28) compared with wild-type cells (unpublished results). This phenotype may be rationalized by a deficiency in the TOR pathway to sense and/or transduce the glucose signal in the absence of ybr077cp, consistent with the profiling data of ybr077cΔ cells (Fig. 4C). We conclude that YBR077C encodes a new factor that partakes in rapamycin-sensitive TOR signaling, and we name it NIR1. It is intriguing that Ybr077cp was detected to bind PI(3,4)P2 in vitro (http://bind.ca), suggesting a possible involvement of PIs in regulating TOR pathway activity (see below and Fig. 4A).
Fig. 4.
Ybr077cp (Nir1p) is a new component of the TOR signaling network. (A) Model of SMIR3 and SMIR4 suppression of the antiproliferative effect of rapamycin possibly by modulating PIs and thereby regulating Ybr077cp activity. (B) Graphic representation of a Ybr077cp protein interaction network and the rapamycin response phenotypes of individual deletion strains. Red, rapamycin-resistant when deleted; green, rapamycin-hypersensitive when deleted; yellow, essential genes whose heterozygous deletion did not produce detectable change in rapamycin sensitivity. (C) Gene-expression evidence for NIR1 function in the Tor network. NC, no change; D, decrease.
Interestingly, in ybr077cΔ cells, SMIR3 is also unable to suppress rapamycin (Fig. 3C), suggesting that SMIR3 may interfere with the same pathway or through similar mechanisms. The lack of additive or synergistic effects between SMIR3 and SMIR4 is in accordance with this possibility. In this light, it is interesting to note that the only strong binder for SMIR3 is Tep1p, whose putative substrates include PI(3,4)P2 based on its homology to PTEN (phosphatase and tensin homologue deleted on chromosome 10) (21, 23). It has been reported that, whereas some PTEN null tumors are resistant (29) to CCI-779, a rapamycin ester currently in clinical trials as an anti-cancer agent, others are more susceptible to it (30, 31). These data suggest the importance of other genetic elements in determining CCI-779 sensitivity (32). Although tep1Δ yeast exhibits normal levels of rapamycin sensitivity, we reasoned that, if Tep1p were indeed involved in TOR signaling, then some of the proteins with which it interacts may also play a role in TOR signaling (“guilt by association”), and their deletion may cause a detectable change in the cell's sensitivity to rapamycin. Tep1p has been found to be associated with six proteins by yeast two-hybrid screening (www.yeastrc.org/cgi-bin/unknown_orfs/orf.cgi?orf=YNL128W#TH). We looked up these proteins in the data set of rapamycin-hypersensitive or -resistant gene deletions determined from a chemical genomic screen (unpublished data). One of the Tep1p-interacting proteins, Apl2p, when deleted conferred hypersensitivity to rapamycin. Apl2p deletion also abolished SMIR3's ability to suppress rapamycin (data not shown). Like Tep1p, Apl2p is highly conserved in mammals by protein sequence comparison. How Tep1p-Apl2p and PIs are involved in TOR signaling and rapamycin sensitivity are fascinating subjects for future investigations.
Our current model for the molecular mechanisms of SMIR3 and SMIR4 in overcoming the antiproliferative effect of rapamycin is depicted in Fig. 4A. From our data, binding of SMIR4 to Ybr077cp apparently causes a gain of function in Ybr077cp. We speculate that SMIR4 by binding to Ybr077cp may dissociate it from a negative regulator (X), thereby allowing cells to grow under compromised TOR function (e.g., by the presence of rapamycin). Database (http://biodata.mshri.on.ca/yeast_grid/servlet/SearchPage) searching suggests the existence of a network of Ybr077cp-interacting proteins (Fig. 4B). Deletion of a few of these proteins conferred opposite response (i.e., resistance) to rapamycin (Fig. 4B) as compared with deletion of Ybr077cp. These proteins and possibly other unidentified interacting proteins (either directly or in a multiprotein complex) represent candidates for the inhibitory activity X proposed in Fig. 4A.
In this study, we report the identification of small molecules that are capable of suppressing rapamycin's inhibitory effects on growth. This chemical genetic approach emulates the logic of classic genetic modifier screening and has led to the discovery of a yeast gene (YBR077C) that appears to take part in TOR signaling. The proteome chip-based target identification strategy provides a systematic and efficient alternative to affinity chromatography, which is biased toward high-abundance proteins. This method also provides valuable proteome-wide information for estimating the specificity and binding affinity of the small molecules.
Supplementary Material
Acknowledgments
We thank T. Mitchison for insightful discussions; P. Nghiem and anonymous reviewers for critical readings of the manuscript and excellent suggestions; J. Broach (Princeton University, Princeton), Y. Jiang (University of Pittsburgh, Pittsburgh), and M. Hall (University of Basel, Basel) for generous providing of yeast strains; M. Luo and R. Cook (Massachusetts Institute of Technology, Cambridge) for assistance in Affymetrix GeneChip hybridizations; S. Hwang and M. Xie for help with experiments shown in Fig. 3B; T. Lorenz and M. Duncan for advice on tetrad analysis; and S. Hwang, M. Duncan, and G. Copeland for helpful comments of the paper. J.H. was a Research Associate and S.L.S. is an Investigator of the Howard Hughes Medical Institute. D.R.S. was supported by a Wellcome Trust Postdoctoral Fellowship. We thank the National Institute for General Medical Sciences for support of this research, the National Cancer Institute (NCI), Merck KGaA, Merck & Co., and the Keck Foundation for support of the Broad Institute Chemical Biology Program, and the NCI for support of the Initiative for Chemical Genetics.
Abbreviations: TOR, target of rapamycin; SMIR, small-molecule inhibitor of rapamycin; YPDA, yeast extract/peptone/dextrose/adenine; PI, phosphoinositide.
References
- 1.DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 2.Hartwell, L. H., Hopfield, J. J., Leibler, S. & Murray, A. W. (1999) Nature 402, C47–C52. [DOI] [PubMed] [Google Scholar]
- 3.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109–126. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, A. C., Ubersax, J. A., Petsch, D. T., Matheos, D. P., Gray, N. S., Blethrow, J., Shimizu, E., Tsien, J. Z., Schultz, P. G., Rose, M. D., et al. (2000) Nature 407, 395–401. [DOI] [PubMed] [Google Scholar]
- 5.Ideker, T., Thorsson, V., Ranish, J. A., Christmas, R., Buhler, J., Eng, J. K., Bumgarner, R., Goodlett, D. R., Aebersold, R. & Hood, L. (2001) Science 292, 929–934. [DOI] [PubMed] [Google Scholar]
- 6.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 7.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 8.Phizicky, E., Bastiaens, P. I., Zhu, H., Snyder, M. & Fields, S. (2003) Nature 422, 208–215. [DOI] [PubMed] [Google Scholar]
- 9.Wohlschlegel, J. A. & Yates, J. R. (2003) Nature 425, 671–672. [DOI] [PubMed] [Google Scholar]
- 10.Mayer, T. U., Kapoor, T. M., Haggarty, S. J., King, R. W., Schreiber, S. L. & Mitchison, T. J. (1999) Science 286, 971–974. [DOI] [PubMed] [Google Scholar]
- 11.Komarov, P. G., Komarova, E. A., Kondratov, R. V., Christov-Tselkov, K., Coon, J. S., Chernov, M. V. & Gudkov, A. V. (1999) Science 285, 1733–1737. [DOI] [PubMed] [Google Scholar]
- 12.Garber, K. (2001) J. Natl. Cancer Inst. 93, 1517–1519. [DOI] [PubMed] [Google Scholar]
- 13.Keith, C. T. & Schreiber, S. L. (1995) Science 270, 50–51. [DOI] [PubMed] [Google Scholar]
- 14.Jacinto, E. & Hall, M. N. (2003) Nat. Rev. Mol. Cell Biol. 4, 117–126. [DOI] [PubMed] [Google Scholar]
- 15.Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. (2001) Science 293, 2101–2105. [DOI] [PubMed] [Google Scholar]
- 16.Thomas, G. & Hall, M. N. (1997) Curr. Opin. Cell Biol. 9, 782–787. [DOI] [PubMed] [Google Scholar]
- 17.Hardwick, J. S., Kuruvilla, F. G., Tong, J. K., Shamji, A. F. & Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. USA 96, 14866–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardenas, M. E., Cutler, N. S., Lorenz, M. C., Di Como, C. J. & Heitman, J. (1999) Genes Dev. 13, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck, T. & Hall, M. N. (1999) Nature 402, 689–692. [DOI] [PubMed] [Google Scholar]
- 20.Peng, T., Golub, T. R. & Sabatini, D. M. (2002) Mol. Cell. Biol. 22, 5575–5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heymont, J., Berenfeld, L., Collins, J., Kaganovich, A., Maynes, B., Moulin, A., Ratskovskaya, I., Poon, P. P., Johnston, G. C., Kamenetsky, M., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 12672–12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maehama, T. & Dixon, J. E. (1998) J. Biol. Chem. 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- 23.Myers, M. P., Pass, I., Batty, I. H., Van der Kaay, J., Stolarov, J. P., Hemmings, B. A., Wigler, M. H., Downes, C. P. & Tonks, N. K. (1998) Proc. Natl. Acad. Sci. USA 95, 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bickle, M., Delley, P. A., Schmidt, A. & Hall, M. N. (1998) EMBO J. 17, 2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmelzle, T., Beck, T., Martin, D. E. & Hall, M. N. (2004) Mol. Cell. Biol. 24, 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heitman, J., Movva, N. R. & Hall, M. N. (1991) Science 253, 905–909. [DOI] [PubMed] [Google Scholar]
- 27.Kim, D. H. & Sabatini, D. M. (2004) Curr. Top. Microbiol. Immunol. 279, 259–270. [DOI] [PubMed] [Google Scholar]
- 28.Granot, D. & Snyder, M. (1991) Proc. Natl. Acad. Sci. USA 88, 5724–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills, G. B., Lu, Y. & Kohn, E. C. (2001) Proc. Natl. Acad. Sci. USA 98, 10031–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neshat, M. S., Mellinghoff, I. K., Tran, C., Stiles, B., Thomas, G., Petersen, R., Frost, P., Gibbons, J. J., Wu, H. & Sawyers, C. L. (2001) Proc. Natl. Acad. Sci. USA 98, 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podsypanina, K., Lee, R. T., Politis, C., Hennessy, I., Crane, A., Puc, J., Neshat, M., Wang, H., Yang, L., Gibbons, J., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10320–10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, S. & Houghton, P. J. (2001) Drug Resist. Update 4, 378–391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.