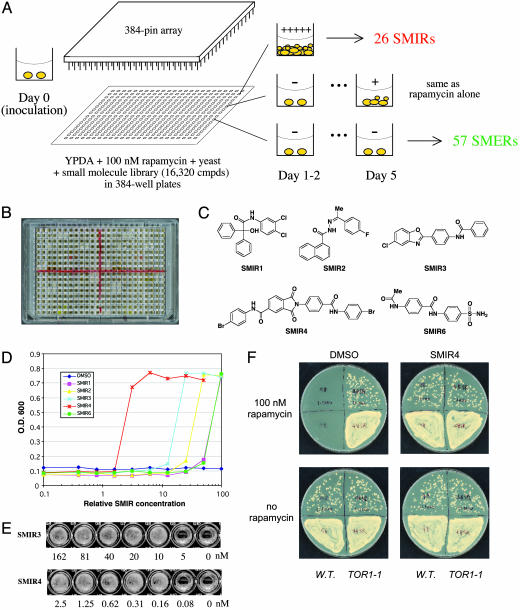
Fig. 1.
A chemical genetic screen for small molecules that modulate rapamycin's antiproliferative effect in yeast. (A) Schematics of the screen. Compounds were transferred from library plates to assay plates (containing growth medium, rapamycin, and yeast cells) by using 384-pin arrays. SMER, small-molecule enhancers of rapamycin. (B) Retest of SMIRs in a 384-well plate. White wells indicate compound-induced yeast growth in the presence of rapamycin; black (transparent) wells indicate no growth. (C) Chemical structures of the fast-acting SMIRs (yeast growth identifiable on day 1, same as the “no rapamycin” control). (D) Dose–response curves for SMIRs in wild-type (rapamycin-sensitive EGY48) cells inoculated in YPDA containing 100 nM rapamycin. (E) Minimal concentrations of SMIR3 and SMIR4 required for yeast growth in YPDA containing 20 nM rapamycin. (F) SMIR4 treatment and the TOR1-1 (S1972R) mutation (26) both confer rapamycin resistance. Cells were plated at two different densities on the upper versus lower halves of the plates (1:1,000).