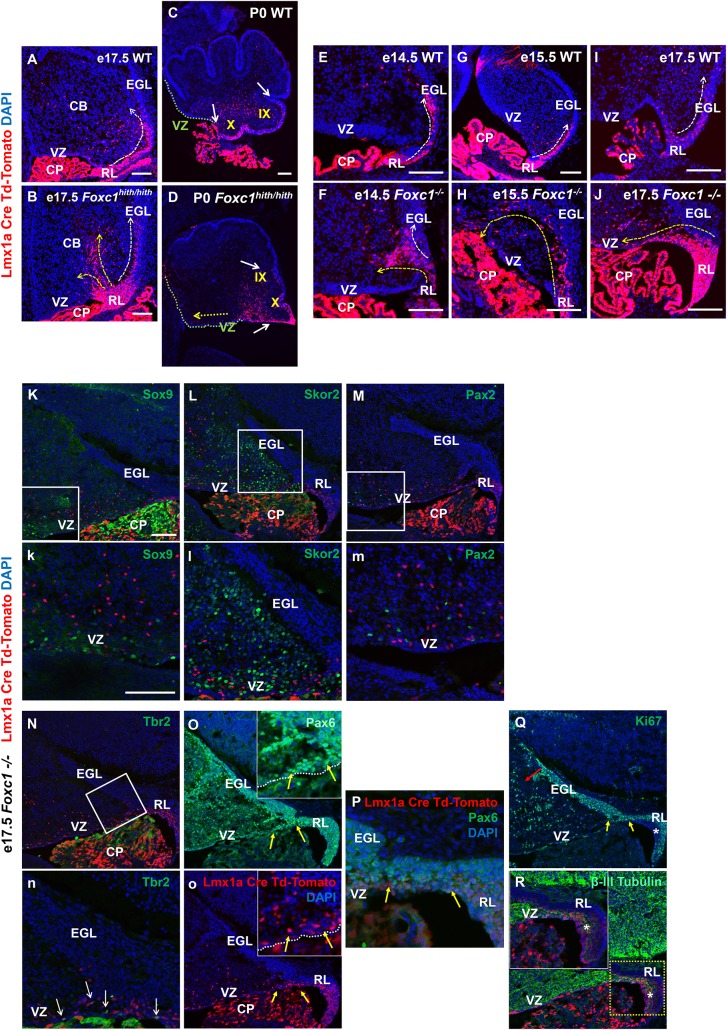
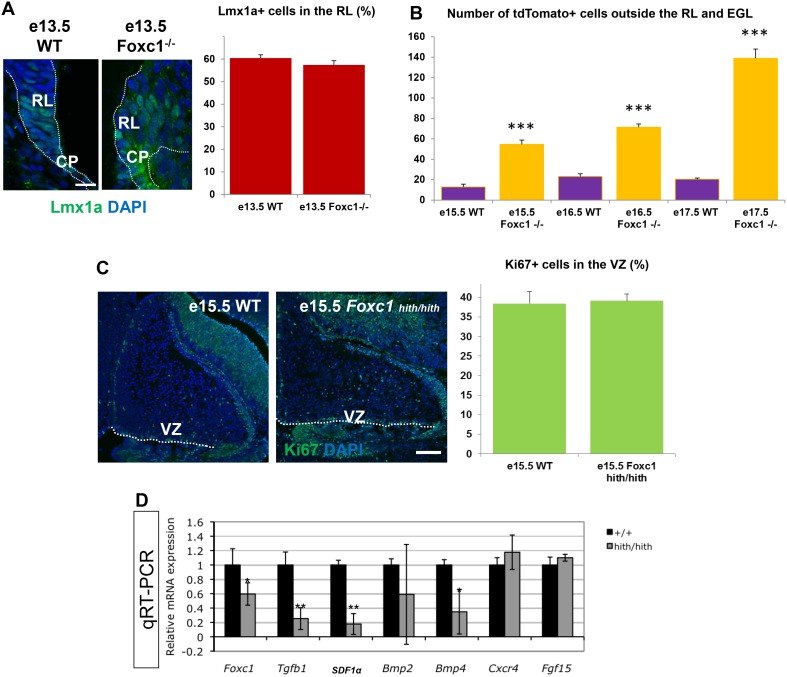
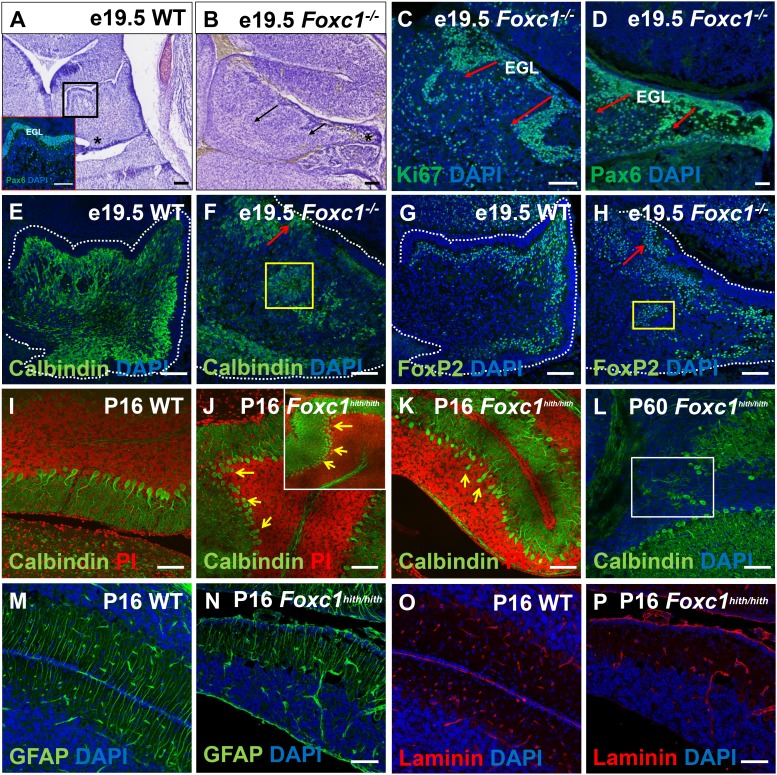
Figure 2. Null and Hypomorphic Foxc1 mutations caused posterior cerebellar foliation defects due to mismigration of cells destined to form the posterior vermis.
(A–J) Lineage analysis of the Lmx1a-cre+ cells in the wild-type mice showed tdTomato expression limited to the RL, EGL and presumptive IGL. Postnatally, fate-mapped cells populated the posterior vermis but did not abut the 2o fissure (C, white arrows). In the wild-type embryonic cerebellum, these cells were present underneath the EGL directly underneath the pial surface (A, E, G, I; white arrow). In Foxc1hith/hith Lmx1a-cre tdTomato mice, cells migrated out of the RL in multiple ectopic streams (B, yellow arrows). Postnatally, in the Foxc1hith/hith mutant cerebellum, ectopic tdTomato+ cells were present along the ventricular surface and the inner cerebellar core (D, yellow arrows). In Foxc1-/- mice (F,H,J), aberrantly migrating Lmx1a-cre tdTomato+ cells were evident by e14.5 in the core (F) and found in the VZ by e15.5 (H, yellow arrow), with an extensive VZ surface presence by e17.5 (J; yellow arrow). Additionally, at e17.5, a large number of fate-mapped mutant cells were abnormally retained in an enlarged RL (J). None of the mutant internal tdTomato+ cells were Sox9+ (K,k), Skor2+ (L,l) or Pax2+ (M,m), and thus had not undergone a VZ lineage fate-switch. A subset of the fate-mapped cells were Tbr2+ (N,n, arrows), as expected of RL-derived unipolar brush cells. All tdTomato+ cells were Pax6+ (O–P). This indicated that they retained their RL origin despite aberrant migration. A subset of the Pax6+ cells is Ki67+ (O, o, Q; yellow arrows) indicating that they retain their ability to divide, while some tdTomato+ cells in the RL (asterisk) are β-III Tubulin+ (R) and Ki67- indicating that they may have differentiated precociously. Scale Bar = 100 µm (A–D, K–Q), 50 µm (E–J).