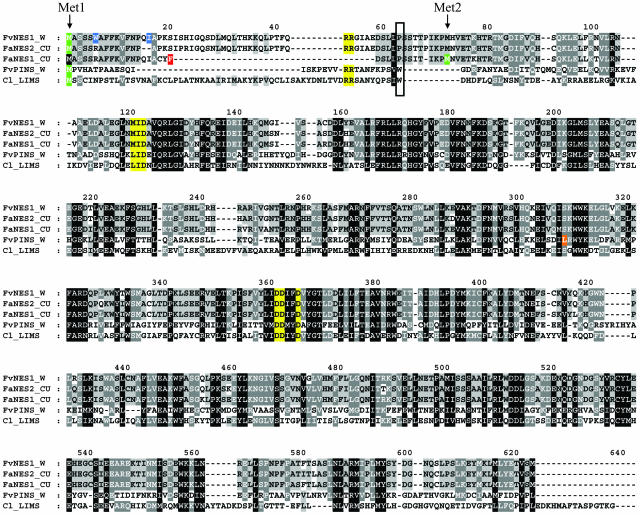
Figure 4.
Protein Sequence Alignment of the Strawberry Terpene Synthases.
The protein sequences of FaNES1, FaNES2, FvNES1, and FvPINS derived from cultivated (CU) and wild (W) strawberry species were aligned to the sequences of the Citrus limon limonene synthase (Cl_LIMS; GenBank accession number AF514287). The stop codon in the FaNES1 gene sequence, between Met1 (shaded green in all sequences except FaNES1) and Met2 (shaded green only in FaNES1) immediately follows the red shaded Phe residue (F). The RR, L/M-I-D, and DDXXD motifs conserved in monoterpene synthases are shaded yellow. Black, gray, and light gray shading represent 100, 80, and 60% conserved identity between residues, respectively. A 2-bp insertion (CC insertion; see also Figure 11) in the cultivated strawberry species results in a frameshift and an immediate stop codon in the middle of the FaPINS gene-coding region (instead of the orange shaded Leu residue [L] in FvPINS). Substitution (W6R) and removal (I16Δ) of the residues shaded blue in FvNES1 resulted in a change of targeting from plastid localization to localization in mitochondria (mainly) as well as to plastids (see also C12 in Figure 9). The residues present at the ninth position succeeding the twin Arg of the RR(x)8W motif normally found in the N-terminal part of class III TPS proteins are boxed. The motif is not entirely conserved in FaNES2 and FvNES1 because it contains a Pro (P) instead of a Trp (W) residue.