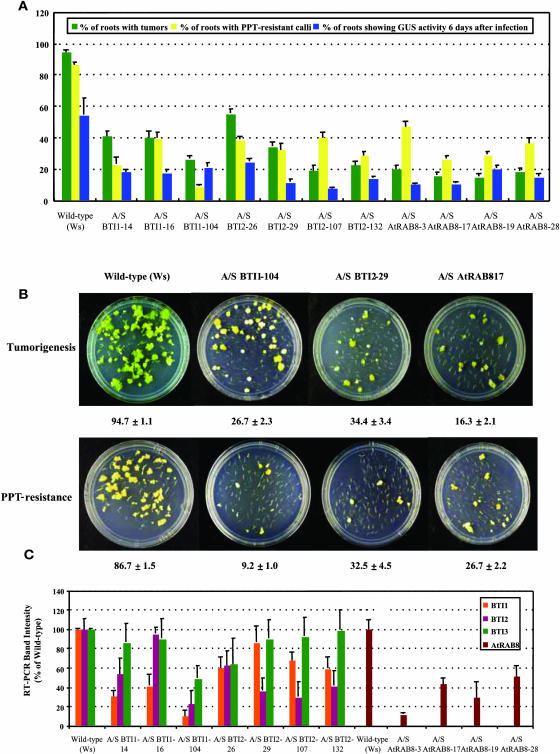
Figure 6.
Transgenic BTI and AtRAB8 Antisense Plants Are Less Susceptible to Agrobacterium-Mediated Root Transformation Than Are Wild-Type Plants.
(A) T2 generation transgenic BTI1, BTI2, and AtRAB8 antisense plants show lower stable and transient transformation efficiencies than do wild-type plants. Green bars represent the percentage of root segments forming tumors 4 weeks after infection with the tumorigenic strain Agrobacterium A208. Yellow bars represent the percentage of root segments forming phosphinothricin-resistance calli 4 weeks after infection with Agrobacterium At872. Blue bars represent the percentage of root segments showing GUS activity 6 d after infection with Agrobacterium At849. At least 20 independent plants were tested for each transgenic line and >80 root segments were examined for each plant. Error bars = se.
(B) Representative plates of BTI1, BTI2, and AtRAB8 A/S transgenic root segments showing reduced frequency of tumor formation and ppt-resistance. The numbers below each plate indicate the average stable transformation efficiency of each line ±se.
(C) Transgenic BTI1 and BTI2 antisense plants show reduced levels of BTI transcripts. Transgenic AtRAB8 antisense (A/S) plants show reduced levels of AtRAB8 transcripts. Transcript levels of each BTI gene and AtRAB8 in A/S transgenic plants are shown as a relative percentage of that of wild-type plants. Data are shown as average values of two RT-PCR reactions from three T2 generation plants of each line. Note that antisense BTI1 and BTI2 plants show a reduced level of BTI2 and BTI1 transcripts, respectively, as well as reduced levels of transcripts targeted by the specific antisense construction. Note also that antisense BTI1 and BTI2 plants generally show a lesser reduction in BTI3 transcripts. Error bars = se.