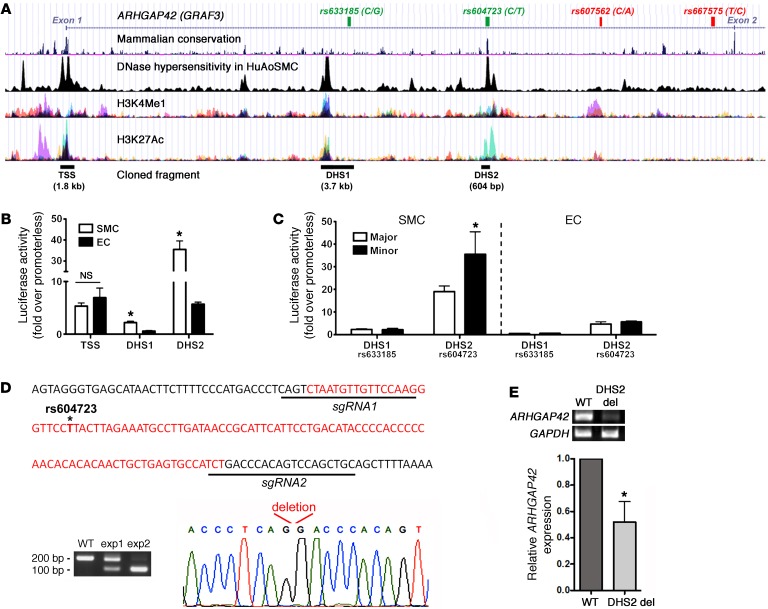
Figure 2. An enhancer within the ARHGAP42 first intron displays strong SMC-specific and allele-specific activity and is required for endogenous ARHGAP42 expression.
(A) Map of the chromatin determinations used to characterize potential regulatory elements near the ARHGAP42 BP-associated locus. The SNPs that define the BP-associated allele (r2 > 0.8) are shown at the top. (B) The indicated DNase-hypersensitive (DHS) regions were cloned into the pGL3 luciferase vector and transfected into primary human bronchial SMCs and mouse ECs. Luciferase activity in cell lysates was measured 2 days later and is expressed as fold over the promoterless pGL3 vector. Data represent mean ± SEM of n = 6 experiments; *P < 0.001 vs. in ECs (Student’s t test). (C) Site-directed mutagenesis was used to test the effects of the major/minor alleles on DHS1 and DHS2 enhancer activity. Data represent mean ± SEM of n = 6 experiments; *P < 0.01 vs. the major allele (Student’s t test). (D) Schematic of the 102-bp deletion (in red) generated by our CRISPR/Cas9–mediated gene editing protocol. (E) ARHGAP42 message was measured by semiquantitative RT-PCR in human bronchial SMC cultures transfected with expression plasmids encoding Cas9 and the guide RNAs shown in D (n = 5). The reduction in ARHGAP42 expression was normalized to the efficiency of DHS2 deletion, which ranged from 45% to 95%. Data represent mean ± SEM of n = 5 separate experiments; *P < 0.05 vs. cells transfected with empty guide RNA expression plasmid (Student’s t test).