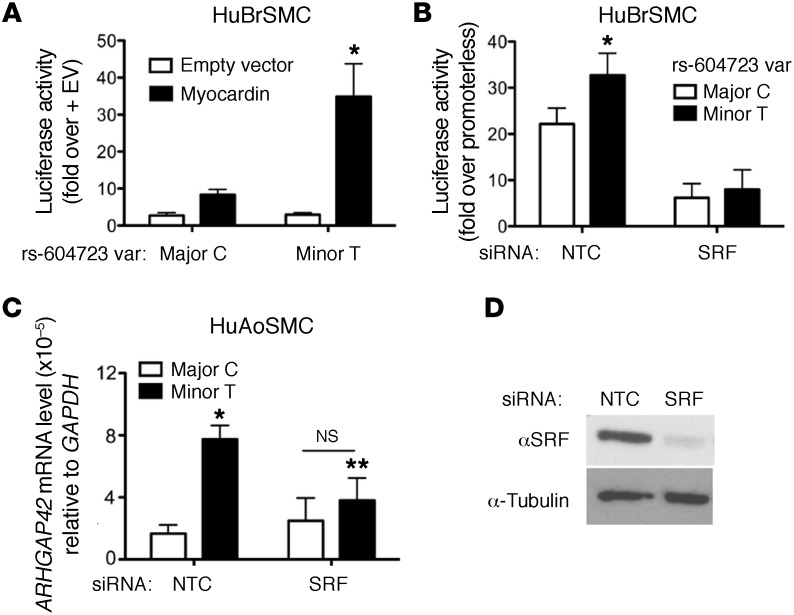
Figure 4. The allele-specific activity of the DHS2 enhancer is SRF-dependent.
(A) Major and minor DHS2 luciferase constructs were transfected into HuBrSMCs along with myocardin or empty expression vector. Data represent mean ± SEM of n = 5 experiments; *P < 0.05 vs. major allele plus myocardin (Student’s t test). (B) DHS2-luciferase activity was measured in HuBrSMCs treated with control (NTC) or SRF siRNA. Data represent mean ± SEM of n = 6 experiments; *P < 0.01 vs. the minor allele (Student’s t test). (C) Allele-specific GRAF3 mRNA levels were measured by semiquantitative RT-PCR in control and SRF knockdown HuAoSMCs. Data represent mean ± SEM of n = 3 experiments; *P < 0.05 vs. the major C allele in control cells; **P < 0.05 vs. minor T allele in control cells (Student’s t test). (D) Confirmation of SRF knockdown in SMCs treated with control or SRF siRNAs. Data are representative of 3 separate experiments.