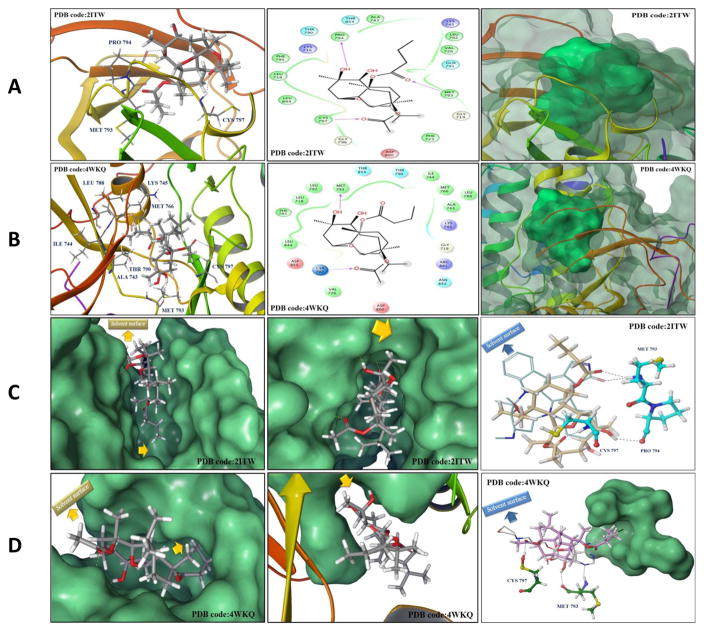
Fig. 11.
(A and B) In silico binding mode of 1 at the ATP binding site of EGFR kinase crystal structures (A) 2ITW and (B) 4WKQ. Left panel; Important interactions of 1 at the EGFR kinase domain, the protein is shown in three-dimensional cartoon presentation. Middle panel; The two-dimensional ligand interaction diagrams (LID) of 1 at the EGFR kinase domain are shown, as generated using the Schrödinger software. Right panel; The transparent protein surface, in aquamarine color, and the solid 1 surface, in spring green color, are shown to emphasize the 1’s shape fitting within the target pocket. (C and D) Left panel; In silico binding pose of 1 at the ATP binding site of EGFR crystal structures (C) 2ITW and (D) 4WKQ, the protein solid surface is shown in aquamarine color with arrows indicating the solvent surface and the deep hydrophobic pocket where the C-11 butyrate group of 1 resides, suggesting the need for bulkier group at this position to improve the 1’s EGFR binding affinity. Middle panel; The hydrophobic subpocket of EGFR crystal structures (C) 2ITW and (D) 4WKQ, where the C-7 methyl group of 1 resides, is indicated by arrows suggesting the need for molecular extension at this position to efficiently fill the EGFR hydrophobic subpocket and improve 1’s EGFR binding affinity. Right panel; Structure overlay for 1 shown in tube with (C) AFN941 and (D) gefitinib conformations shown in thin tube, as obtained from EGFR crystal structures 2ITW and 4WKQ, respectively, via docking simulations. The important HB interactions exerted by 1 and the original ligands towards critical amino acids within the EGFR kinase pocket are also shown. In particular, 1 engaged in a crucial HB with Cys797, conferring T790M EGFR mutant inhibitory activity, unlike the hexahydroindole ring of AFN941 and the propylmorpholino group of gefitinib, which missed this interaction. The protein solid surface for the deep hydrophobic pocket where the C- 11 butyrate of 1 perfectly overlaid with the aniline ring of gefitinib at the back of the ATP-binding cleft is also shown in aquamarine color and the solvent surface is indicated in arrows.
Abbreviations: ATCC, American type culture collection; CD30, cluster of differentiation 30 a type I transmembrane glycoprotein belonging to the TNF receptor superfamily; CD31, platelet endothelial cell adhesion molecule 1; EGF, epidermal growth factor; EPHB4, ephrin type-B receptor 4; ERK, extracellular signal-regulated kinase; FGFR, fibroblast growth factor receptor; FLT3, fms-related tyrosine kinase 3; JAK2, janus kinase 2; L858R, leucine 858 mutated to arginine; L861Q, leucine 861 mutated to glutamine; M, mitosis; MTT, 3-[4,5-dimethylthiazol-2- yl]-2,5-diphenyltetrazolium bromide; PDB, protein data bank; PLK1, polo-like kinase 1; P70S6K1, phosphorylation of 70-kDa ribosomal protein S6 kinase 1; RIPA, radioimmunoprecipitation assay; RMSD, root mean square displacement; RPMI, Roswell park memorial institute; TBST, tris-buffered saline with Tween 20; TIE2, tyrosine kinase with immunoglobulin-like and EGF-like domains 2; T790M, threonine 790 mutated to methionine; TNBC, triple negative breast cancer; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; Wnt, wingless-type MMTV integration site family member.