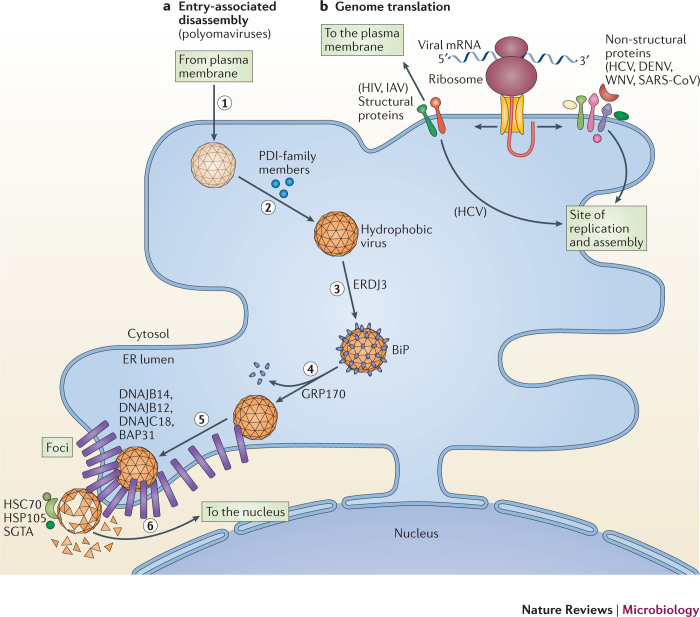
Figure 2. Exploiting the ER during the early stages of infection.
a | Entry-associated disassembly. Viruses must disassemble their capsid to release their genome. Members of the Polyomaviridae disassemble their capsid by co-opting components of the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway. To cause infection, polyomaviruses undergo receptor-mediated endocytosis and traffic to the ER (step 1). Once at the ER, they use protein disulfide isomerase (PDI)-family members to isomerize and reduce viral disulfide bonds (step 2). These events partially disassemble the virus particles to form hydrophobic viruses that engage binding immunoglobulin protein (BiP) through the activity of ER DNAJ domain-containing protein 3 (ERDJ3; step 3). 170 kDa glucose-regulated protein (GRP170) then releases polyomavirus from BiP, enabling the hydrophobic virus to insert into the ER membrane (step 4). The membrane-inserted virus reorganizes selective ER membrane proteins (B cell receptor-associated protein 31 (BAP31) and the J proteins DNAJ homologue subfamily B member 12 (DNAJB12), DNAJB14 and DNAJ C18) in the lipid bilayer to form foci (step 5). The membrane-attached cytosolic extraction machinery (heat shock cognate protein 70 (HSC70)–heat shock protein 105 (HSP105)–small glutamine-rich tetratricopeptide repeat-containing protein-α (SGTA)) then ejects the virus from the foci into the cytosol in a reaction that simultaneously disassembles the virus (step 6). Cytosolic disassembly enables the resulting core virus particle to move into the nucleus to cause infection. b | Genome translation. Some viruses exploit the ER-associated biosynthetic machinery to translate their genetic code in two ways: the translation of viral structural proteins that are incorporated into virions (for example, the envelope glycoprotein of HIV and haemagglutinin of influenza A virus (IAV)) and the translation of viral non-structural proteins that promote the subsequent viral replication step. This is evident during translation of the positive-sense RNA ((+)RNA) genome of viruses in the Flaviviridae (hepatitis C virus (HCV), dengue virus (DENV) and West Nile virus WNV) and Coronaviridae (severe acute respiratory syndrome coronavirus (SARS-CoV)) families, in which the newly synthesized replication proteins target to sites of virus replication on the ER (or ER-derived) membrane compartments in preparation for replication. At these sites, the physical architecture of the ER membrane effectively acts as a scaffold to recruit the viral replication proteins.