Fig. 2.
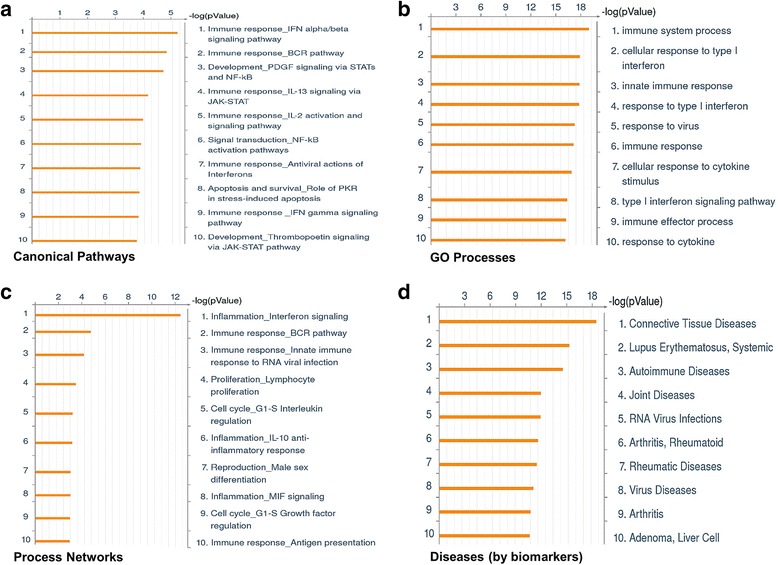
Functional annotation and pathway-based analysis of VL-blood associated transcriptional profile. Gene expression values from peripheral blood samples of 8 VL patients and 6 healthy individuals were compared, generating a trimmed list of 99 DEGs (4 up-and 91 down-regulated). Algorithms use the uploaded VL-blood associated DEGs as the input list and map each gene to a network object in the MetaCore database. Pathway and network analysis of the VL-blood transcriptional profile demonstrate an enrichment of the following categories: (a) canonical pathways (b) GO processes (c) process networks and (d) disease by biomarkers. Top ten enrichments are sorted and ranked by p-value shown on a logarithmic scale. A lower p-value indicates higher statistical significance
