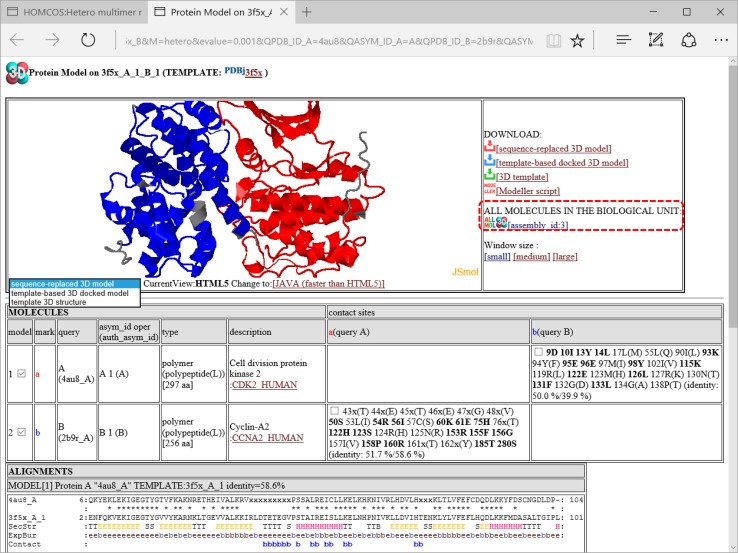
Fig. 11.
A snapshot of the window for 3D model. This page shows a 3D model of the complex of CDK5 (PDBcode:4au8 chainA) and cyclin B1(PDBcode:2b9r chain A) modeled by the template complex of CDK2 and cyclin A2 (PDBcode:1h27). The 3D structure of the sequence-replaced model is shown at the top left of the page using JSmol or Jmol. This model is created simply by replacing the residue name and number with those of the query protein according to the BLAST alignment. Non-aligned regions of the template structure are displayed in gray color. At the top right of the page, there are several links for downloading models are placed: downloading sequence-replaced model, template-based docked 3D model, template, and MODELLER script [53]. The link “ALL MOLECULES IN THE BIOLOGICAL UNIT” generates a new page including all of the molecules in one of the biological units (shown in Fig. 12). At the middle of the page, all of the template molecules in this model are shown. Residue numbers and residue names of the contact sites of the query proteins are shown in the left column (“contact sites”). If the site has the common amino acid for the query and the template, the site is shown in bold. At the end of the column “contact sites”, two sequence identities are shown, such as “50.0/39.9” and “51.7/58.6”. The first number is the sequence identity of the contact site, and the second number is the identity of all the aligned sites. The identity of the contact site is a good measure for the quality of the model, especially for the small chemical compound. At the bottom of the page, alignments between the query and the template proteins are shown. Contact sites are indicated by the “mark” letters, such as “b”