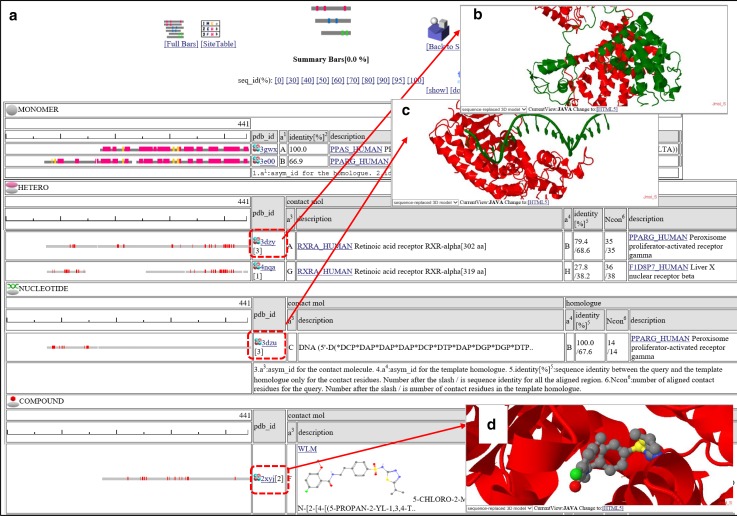
Fig. 5.
Snapshots of the contact “Summary Bars” view of “searching contact molecules for query protein”. The amino acid sequence of human PPAR-delta (PPARD_HUMAN) is used as the query. a The Summary Bars view. Bars show regions of aligned homologous structure. The width of the bar corresponds to the length of the query protein. In the top “MONOMER” table, the secondary structures are shown by colored bars (red: α-helices, yellow: β-strands). In the other tables (“HETERO”, “NUCLEOTIDE”, and “COMPOUND”), contacting residues with other molecules are shown in small red boxes. If a user clicks the “3D” icon, then the 3D model view windows appears. In the column “identity[%]”, two sequence identities are shown, such as “79.4/68.6” and “100.0/67.6”. The first number is the sequence identity of the contact site, and the second number is the identity of all the aligned sites. The identity of the contact site is a good measure for the quality of the model, especially for the small chemical compound. b A 3D model of a hetero protein complex, based on the template 3dzy. The contact protein is RXRA_HUMAN (asym_id = A), and the homologue is PPARG_HUMAN (asym_id = B; sequence identity = 68.6 %). c A 3D model of a protein-nucleotide complex based on the template 3dzu. The contact molecule is the DNA single strand (asym_id = C). The homologue is PPARG_HUMAN (asym_id = B; sequence identity = 67.6 %). The figure shows one protein – single stranded DNA complex; however, the biological unit of 3e00 is composed of two protein chains and double stranded DNA. To display all of the molecules in the biological unit, click the icon “ALLMOL” in the 3D model view page (see Figs. 11, 12). d A 3D model of a protein-compound complex based on the template 2xyj. The contact molecule is WLM (asym_id = F). The homologue is PPARD_HUMAN (asym_id = A; sequence identity = 100 %). This complex is an experimentally determined 3D structure for the query complex, rather than a prediction