Figure 2.
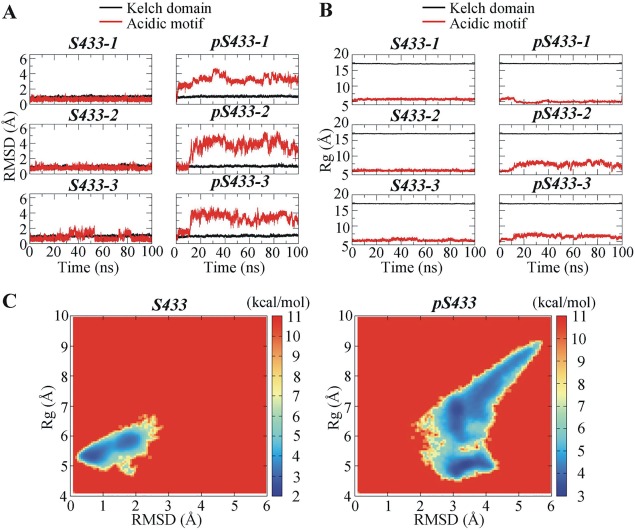
Time‐dependent structural alterations of the Kelch domain of KLHL3 and the acidic motif (AM) of WNK4. (A) Root mean square deviation (RMSD) as a function of simulation time. (B) Radius of gyration (Rg) as a function of simulation time; (C) Free energy landscape for the AM of WNK4 with respect to RMSD and Rg. In (A) and (B), three independent simulations for the Kelch domain−AM system containing unphosphorylated S433 (S433‐1 to S433‐3) and phosphorylated S433 (p433‐1 to pS433‐3) were performed.
