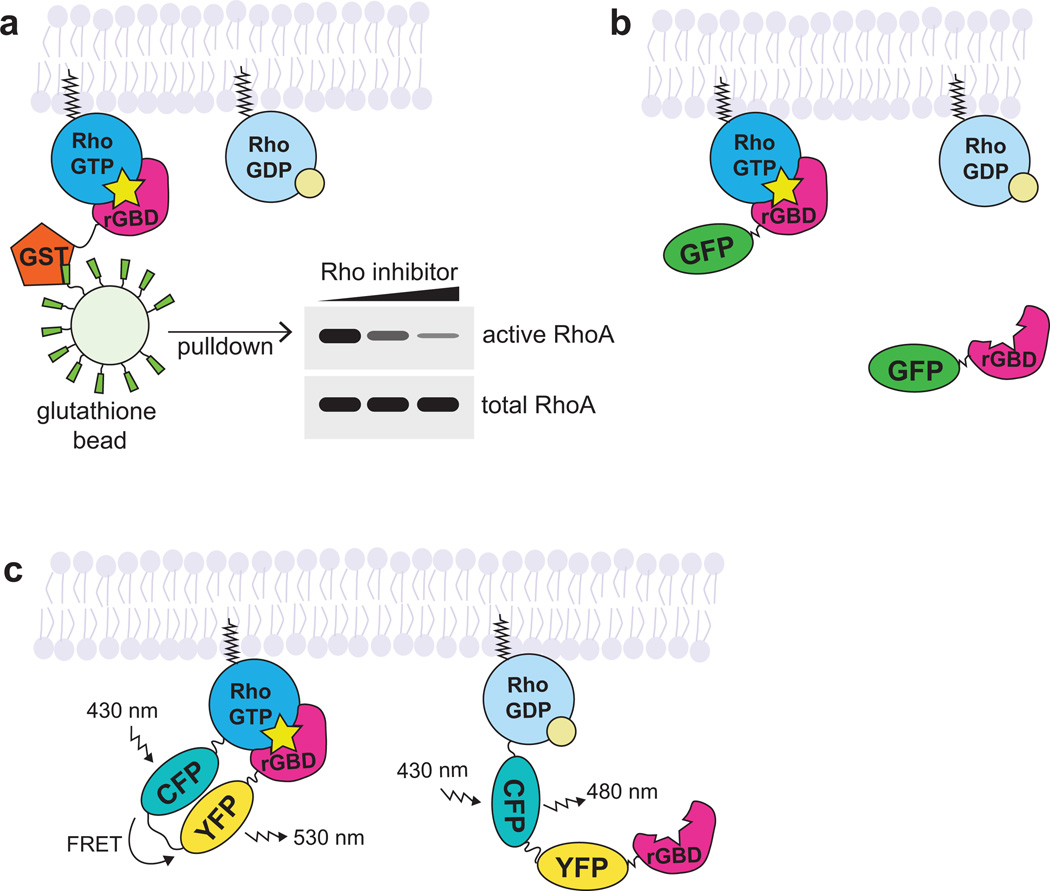
Figure 2. Approaches for studying active Rho GTPases.
a. In the GBD affinity pull-down approach, the active GTPase is pulled down with a GST-tagged effector GBD specific for the GTPase of interest (rGBD = Rhotekin GBD, which binds RhoA, B, and C). The amount of GTPase in the pull-down is compared with total GTPase in the sample to approximate the pool of active GTPase in the sample.
b. In the effector translocation (GBD probe) approach, the effector GBD is fluorescently-tagged (with GFP) and binds to the endogenous active GTPase. Local increase in fluorescence intensity over background is interpreted as increased active GTPase.
c. In the GTPase-effector FRET biosensor (FRET biosensor) approach, the GTPase is tagged with a donor fluorophore (CFP) and the effector GBD is tagged with an acceptor fluorophore (YFP). A unimolecular Rho biosensor is shown here. When the GTPase is inactive, the donor fluorophore emits light. When the GTPase is active, the donor fluorophore excites the acceptor fluorophore.