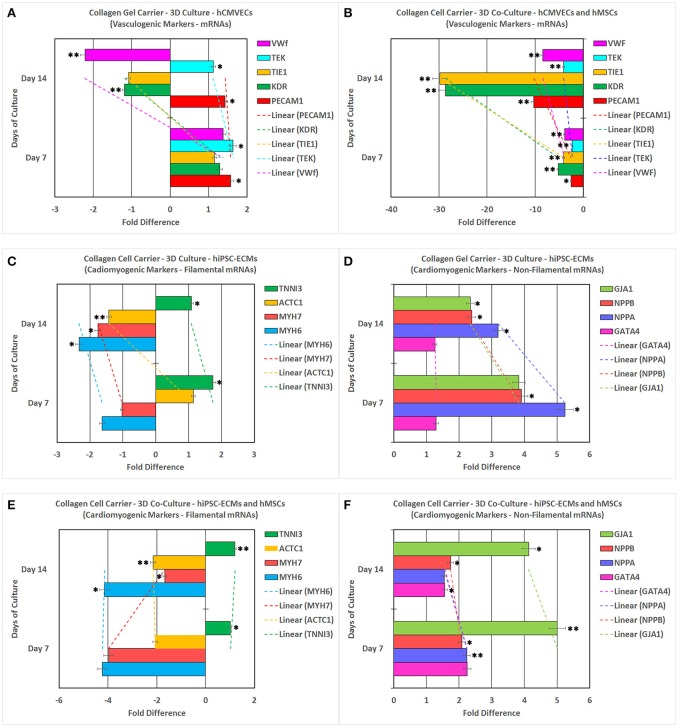
Figure 11.
Reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR) analysis of various key vasculogenic and cardiomyogenic markers. PECAM1 (platelet and endothelial cell adhesion molecule 1), KDR (kinase insert domain receptor, a type III receptor tyrosine kinase), TIE1 (tyrosine kinase with immunoglobulin-like and EGF-like domains 1), TEK (TEK tyrosine kinase, endothelial), and VWF (von Willebrand factor) expression (abscissa) as a function of time (ordinate). hCMVECs cultured onto CCCs in complete microvascular endothelial cell growth medium (A). hCMVECs/hMSCs co-cultured onto CCCs in complete microvascular growth medium (B). Similarly, MYH6 (myosin, heavy chain 6, cardiac muscle, alpha), MYH7 (myosin, heavy chain 7, cardiac muscle, beta), ACTC1 (actin, alpha, cardiac muscle 1), TNNI3 (troponin I3, cardiac type), GATA4 (GATA binding protein 4), NPPA (natriuretic peptide A), NPPB (natriuretic peptide B), and GJA1 (gap junction protein, alpha 1) expression (abscissa) as a function of time (ordinate). hiPSC-ECMs cultured onto CCCs in complete myocyte medium (C,D). hiPSC-ECMs/hMSCs co-cultured onto CCCs in complete myocyte medium (E,F). The calibrator control included hCMVECs day 0 sample for vasculogenic cultures and hiPSC-ECMs day 0 sample for cardiomyogenic cultures, and the target gene expression was normalized by three non-regulated reference gene expressions, viz., GAPDH, β-ACTIN, and either G6PD or RPLP0. The expression ratio (abscissa) was calculated using the Relative Expression Software Tool–384 (REST-384 - version 2). The values were means ± standard errors for three independent cultures (n = 3), *p < 0.05; **p < 0.001.