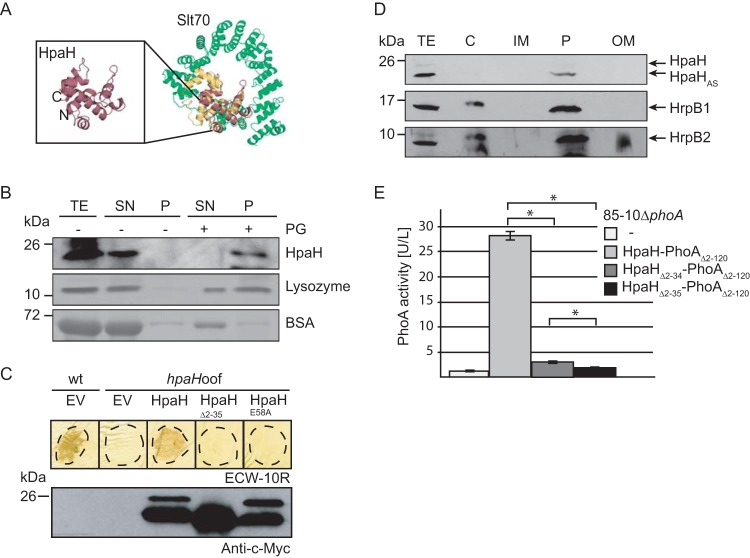
FIG 2.
HpaH localizes to the bacterial periplasm and interacts with peptidoglycan. (A) HpaH shares predicted structural similarity with the LT Slt70 of E. coli. The structure of HpaH was predicted by PHYRE (http://www.sbg.bio.ic.ac.uk/phyre) and aligned with the structure of Slt70 from E. coli (UniProtKB/Swiss-Prot accession number P0AGC3.1) using the PyMOL software (http://www.pymol.org/). HpaH is shown in red, the C-terminal domain of Slt70, which contains the active site (30), in yellow, and additional Slt70 protein regions in green. The N and C termini of HpaH are indicated. (B) HpaH binds to peptidoglycan in vitro. HpaH, lysozyme, and BSA were incubated in the absence (−) or presence (+) of peptidoglycan (PG). Total cell extracts (TE) as well as proteins of the supernatant (SN) and the pellet (P) fractions were analyzed by SDS-PAGE and Coomassie blue staining or immunoblotting using antibodies specific for HpaH. (C) Mutation of the predicted catalytic glutamate residue at position 58 abolishes HpaH function. Strains 85-10 (wt) and 85-10hpaHoof (hpaHoof) containing the vector pBRM (EV) or HpaH derivatives as indicated were infiltrated at a density of 5 × 107 CFU ml−1 into leaves of resistant ECW-10R pepper plants. Leaves were destained in ethanol at 2 dpi. Dashed lines mark the infiltrated areas. For protein analysis, equal amounts of cell extracts (adjusted according to the cell density) were analyzed by immunoblotting using a c-Myc epitope-specific antibody. (D) Fractionation studies with HpaH. Strain 85*hpaHoof containing HpaH–c-Myc was incubated in secretion medium. Fractions enriched in the cytoplasm (C), the inner membrane (IM), the periplasm (P), and the outer membrane (OM) were separated by ultracentrifugation and analyzed by immunoblotting using a c-Myc epitope-specific antibody. Blots were reprobed with antibodies specific for HrpB1 and HrpB2. (E) HpaH-PhoAΔ2–120 displays phosphatase activity. Strain 85-10ΔphoA without an expression construct or containing HpaH-PhoAΔ2–120 fusions as indicated was incubated in secretion medium. Phosphatase activities were determined using pNPP as the substrate. All fusion proteins were stably synthesized, as shown in Fig. S3 in the supplemental material. Values represent the means of three measurements of one strain. Error bars represent standard deviations, and asterisks denote statistically significant differences according to the Student t test (P > 0.001).