Fig. 3.
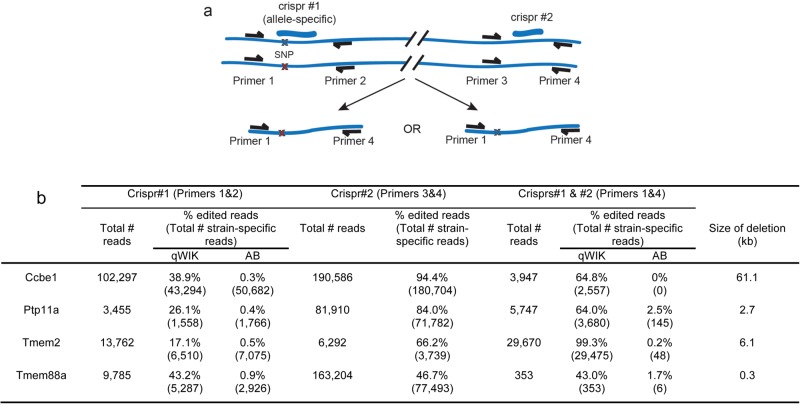
Chromosome-specific deletions created using CRISPR/Cas9 allelic bias. (A) Schematic representation of experiment generating genomic deletions using two CRISPR/Cas9 gRNAs, where ‘crispr#1’ targets a polymorphic site and ‘crispr#2’ targets a non-polymorphic site. Blue crosses indicate qWIK-specific SNP. Red crosses indicate AB-specific SNP. (B) Table depicting the editing efficiency for the various gRNAs injected in isolation or in combination and quantified by sequenced PCR products spanning each edited site alone or the deleted regions. Total reads represents all mapped reads (qWIK-specific, AB-specific and unequivocal). Percentage of edited reads for crispr#1 only (primers 1&2) and #2 only (primers 3&4) expressed as a function of total number of strain-specific reads. Percentage of edited reads for crisprs#1 and #2 (primers 1&4) is calculated as a function of total read number.