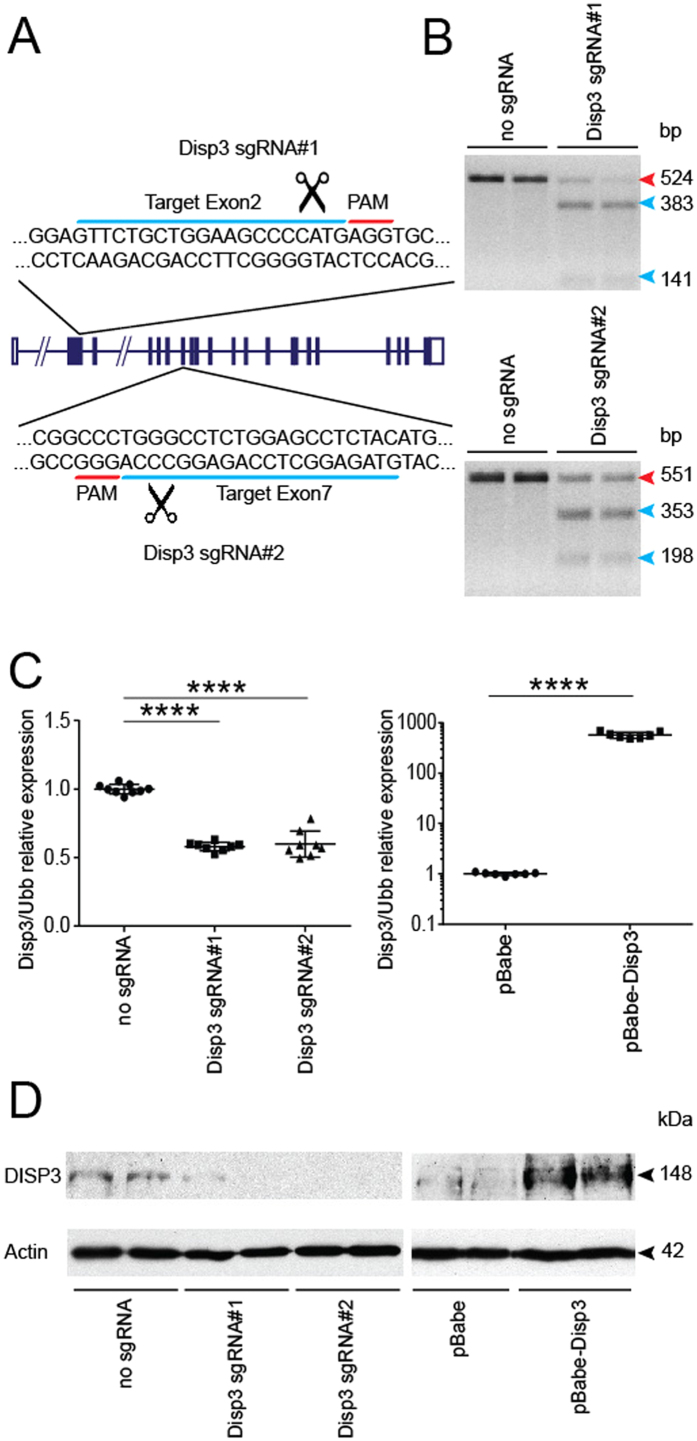
Figure 2. Modulation of DISP3 levels in NS-5 cells.
(A) A scheme of the Disp3 gene showing two sgRNA target sites for Disp3 targeting via RNA-guided CRISPR/Cas9 endonuclease. The 20 nt target sequence (blue) is represented along with the neighboring PAM motif (red). (B) T7 endonuclease I assays confirm indels induced by Disp3 sgRNA#1 and Disp3 sgRNA#2 in two independent cell batches. The red arrow indicates the wild-type DNA fragment, blue arrows indicate mutant DNA fragments. (C) Quantitative RT-PCR analysis of Disp3 mRNA. Cells infected with a lentivirus encoding Cas9 and Disp3 sgRNA#1 or Disp3 sgRNA#2 and control without sgRNA (left scatter plot); cells infected with a retrovirus encoding human DISP3 (pBabe-Disp3) and control empty vector (pBabe) (right scatter plot). Ubb was used as a reference gene. Data represent the mean of biological replicates with error bars indicating standard deviation and the level of statistical significance (****P < 0.0001). Mean values: no sgRNA = 1.00, Disp3 sgRNA#1 = 0.58, Disp3 sgRNA#2 = 0.60, pBabe = 1.00, pBabe-Disp3 = 576. (D) Level of DISP3 protein confirmed by western blot. Actin served as a loading control.