Abstract
During meiosis, a specialized chromosome structure is assembled to promote pairing/synapsis of homologous chromosomes and meiotic recombination, a process yielding chiasmata between homologs to ensure accurate segregation. Meiosis‐specific cohesin complexes mediating sister chromatid cohesion play pivotal roles in almost all these events, including synaptonemal complex (SC) formation. In this issue of EMBO Reports, Agostinho and colleagues have examined chromosome axes and SC structures by taking advantage of a hypomorphic Stag3 mutant in which the levels of the cohesin subunit REC8 are partly reduced 6. Using super‐resolution microscopy, the authors illuminate previously unforeseen chromosome axis structures, showing locally separated axes in regions where REC8 is absent, regardless of RAD21L or RAD21 cohesin localization. Furthermore, they assessed the relationship between sister chromatid cohesion and inter‐sister SC formation, demonstrating that “axial opening” in the REC8‐free region is accompanied by illegitimate SC formation between sister chromatids. This study highlights the physiological importance of REC8 in sister chromatid cohesion and proper SC formation during meiosis, suggesting a new model in which a high density of REC8 deposition along the chromosome prevents illegitimate inter‐sister SC formation.
Subject Categories: Cell Cycle
When chromosomes replicate in S‐phase, sister chromatids are held together by a mechanism termed sister chromatid cohesion that enables accurate chromosome segregation in both mitosis and meiosis. In somatic cells, sister chromatid cohesion is mediated by the cohesin multi‐protein complex, which is comprised of four core subunits: SMC1α and SMC3, the kleisin family protein RAD21, and either one of the accessory subunits SA1 and SA2. In mammalian germ cells, the cohesin complex contains in addition to RAD21 the meiosis‐specific kleisin subunits REC8 and RAD21L 1, 2, 3 (Fig 1A). Furthermore, SMC1α and SA1/SA2 are largely replaced by other meiosis‐specific cohesin subunits, SMC1β and STAG3, respectively 4, 5. Given that STAG3, SMC1β, and SMC3 are commonly shared in these meiotic cohesin complexes, it is assumed that the kleisin subunits REC8, RAD21L, and RAD21 have specific roles in the cohesin complexes during meiosis 1, 2.
Figure 1. Synaptonemal complex (SC) assembly in the cohesin mutant mice.
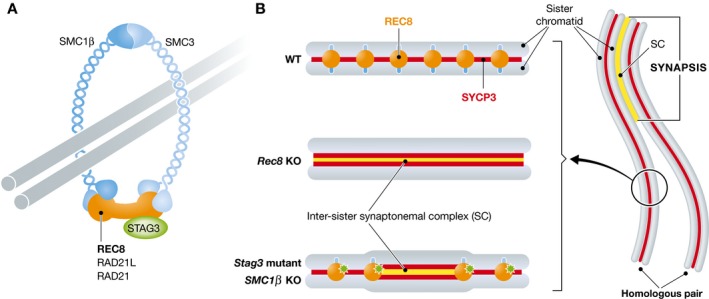
(A) The meiosis‐specific cohesin complex contains four core subunits, SMC1β, SMC3, STAG3, and one of the kleisin subunits REC8, RAD21L, or RAD21. (B) In wild‐type cells, the chromosome axis in the unsynapsed region is recognized as a single line of SYCP3‐labeled structures along which a high density of REC8 foci can be detected. In Rec8 KO spermatocytes, the axis of the univalent chromosome is resolved as two separate SYCP3‐labeled structures. In Stag3 mutant and Smc1β KO cells, the chromosome axis is regionally separated in those areas where REC8 is absent. The “axial opening” is flanked by REC8 and is accompanied by inter‐sister SC formation.
In the meiotic prophase, REC8 is localized along chromosomes before meiotic DNA replication and persists throughout the first meiotic division and, at least at centromeres, until metaphase II. In contrast, RAD21L transiently appears on the chromosomes after DNA replication and culminates at the leptotene/zygotene stage. While RAD21 is detectable in testicular mitotic cells, it is not detected in nuclei from early leptotene until zygotene, in contrast to the patterns of RAD21L and REC8. Thus, the kleisin subunits endow distinct cohesin complexes with different spatiotemporal localization patterns on chromosomes during meiotic prophase. Accumulating evidence suggests that meiosis‐specific cohesin complexes play essential roles not only in sister chromatid cohesion, but also in SC formation. However, it is largely elusive how distinct cohesin complexes exert their specific functions in sister chromatid cohesion and SC formation.
Using super‐resolution microscopic analyses (SIM and STED), Agostinho and colleagues have now examined chromosome axis and SC structures in mouse mutants defective in various cohesin subunits (Rec8, Stag3, or Smc1β) 6. In wild‐type spermatocytes, the chromosome axis, which consists of two sister chromatids, is labeled by a single line of the axial element (AE) component SYCP3. Strikingly, however, the entire axis of univalent chromosomes can be resolved as two separate SYCP3‐labeled structures in Rec8 knockout (KO) cells, which is consistent with previous electron microscopic observations 7, 8. Moreover, similar to these observations in Rec8 KO spermatocytes, the chromosome axis is also regionally separated in Stag3 mutant and Smc1β KO cells, which has previously not been observed, due to the low resolution of ordinary optical microscopic tools (Fig 1B).
Notably, the inter‐axis distance at these “axial opening” regions in Stag3 mutant and Smc1β KO cells is comparable to that of synapsed homologous chromosomes in the wild type (150–175 nm). Previous reports have indicated that illegitimate SCs might be assembled between sister chromatids in Rec8 KO cells 9, 7. Similarly, the authors detected SC components (SYCP1, SYCE1, SYCE2, and TEX12) at these local “axial opening” regions in the Stag3 mutant and Smc1β KO cells. This suggests that tripartite SC assembly, which is discernible from that between synapsed homologs in the wild type, occurs in local “axial opening” regions in the absence or at reduced levels of particular cohesin subunits (Fig 1B).
Previously, the same group showed that Stag3 mutant spermatocytes exhibit a hypomorphic phenotype during the progression of meiotic prophase, in which the REC8 cohesin level is partly reduced 5. This evidence prompted the authors to assess the distribution of REC8 at local “axial opening” regions in the Stag3 mutant. Intriguingly, in this mutant, REC8 is absent at local “axial opening” regions, while REC8 foci are detected between tightly associated sister chromatid axes. Notably, local “axial opening” regions are flanked by REC8 foci, suggesting that REC8 defines the borders of these regions. Importantly, the REC8‐free chromosome axis shows regional “axial opening”, despite the fully localized RAD21L cohesin or partially localized RAD21 cohesin components along the two sister chromatid axes. Furthermore, the authors demonstrate that these phenomena are also present in Smc1β KO cells. Thus, REC8, but not RAD21L or RAD21, is required to sustain the regional “axial opening” and prevent illegitimate SC assembly between sister chromatids. The super‐resolution microscopic analyses clearly demonstrate that in wild‐type spermatocytes, REC8 foci are detected within short intervals along yet‐synapsed autosomes without any detectable illegitimate inter‐sister SC assembly. In contrast, in Stag3 mutant cells, REC8 foci are detected at longer intervals along the chromosome axis with illegitimate inter‐sister SC formation. These data lead the authors to conclude that the distribution of a high density of REC8 foci along the chromosome axis at short intervals constrains the sister chromatid axes and prevents illegitimate inter‐sister SC assembly (Fig 1B).
A previous study showed that sister chromatid cohesion is maintained solely depending on REC8 at least in bivalent metaphase chromosomes 10. However, the current study raises interesting questions as to how individual meiosis‐specific cohesion complexes contribute to sister chromatid cohesion and chromosome axis integrity in prophase I. The authors propose local “axial opening” at regions locally devoid of REC8‐mediated sister chromatid cohesion in the Stag3 and Smc1β mutants, which is always accompanied by illegitimate inter‐sister SC formation. Nevertheless, the causal relationship between inter‐sister SC assembly and local “axial opening” still remains elusive. It is possible that ectopic inter‐sister SC assembly forces sister chromatid axes to be separated, pushing away REC8 cohesin to the flanking regions, given that cohesin rings can slide along the chromosome. Further, inter‐sister SC formation in Rec8 KO largely depends on SPO11, which introduces DNA double‐strand breaks 8. Thus, it is tenable that the atypical cohesin RAD21L establishes sister chromatid cohesion depending on DNA double‐strand breaks as reported in mitotic yeast cells. However, further investigations are required to clearly establish the division of labor between REC8 and RAD21L.
See also: A Agostinho et al (June 2016)
References
- 1. Lee J, Hirano T (2011) J Cell Biol 192: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ishiguro K, Kim J, Fujiyama‐Nakamura S et al (2011) EMBO Rep 12: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Llano E, Herran Y, Garcia‐Tunon I et al (2012) J Cell Biol 197: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Revenkova E, Eijpe M, Heyting C et al (2004) Nat Cell Biol 6: 555–562 [DOI] [PubMed] [Google Scholar]
- 5. Fukuda T, Fukuda N, Agostinho A et al (2014) EMBO J 33: 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agostinho A, Manneberg O, van Schendel R et al (2016) EMBO Rep 17: 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu H, Beasley MD, Warren WD et al (2005) Dev Cell 8: 949–961 [DOI] [PubMed] [Google Scholar]
- 8. Ishiguro K, Kim J, Shibuya H et al (2014) Genes Dev 28: 594–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bannister LA, Reinholdt LG, Munroe RJ et al (2004) Genesis 40: 184–194 [DOI] [PubMed] [Google Scholar]
- 10. Tachibana‐Konwalski K, Godwin J, van der Weyden L et al (2010) Genes Dev 24: 2505–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]


