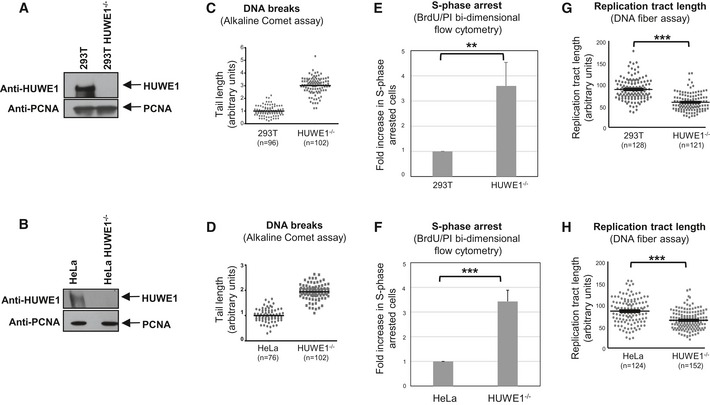
Figure 1. HUWE1‐knockout cells show genomic instability and increased replication stress.
-
A, BWestern blot showing the absence of HUWE1 protein in 293T (A) and HeLa (B) cells subjected to CRISPR/Cas9‐mediated HUWE1 deletion.
-
C, DHUWE1‐knockout 293T (C) and HeLa (D) cells show increased DNA breaks in the absence of exogenous DNA damage treatment. Results from the alkaline comet assay are shown. The “n” numbers of comet tails analyzed (pooled from two independent experiments), as well as the mean, are indicated on the graphs. HUWE1‐knockout HeLa cells did not show increased breakage in the neutral comet assay (Fig EV1B), indicating that the majority of breaks observed in cycling HUWE1‐knockout cells are not double‐strand breaks, but rather single‐strand breaks and other types of lesions.
-
E, FIncreased S‐phase arrest in HUWE1‐knockout 293T (E) and HeLa (F) cells. Cycling cells were incubated with BrdU and subjected to BrdU/PI bi‐dimensional flow cytometry. Representative flow cytometry profiles are presented in Fig EV1D–F. Bars represent the fold increase in the percentage of cells with S‐phase DNA content (between 2N and 4N) but negative for BrdU staining. Bars represent the average of three independent experiments, with error bars showing SD. The P‐values are 0.0091 (E) and 0.0007 (F).
-
G, HThe DNA fiber assay shows reduced replication tract length in HUWE1‐knockout 293T (G) and HeLa (H) cells in the absence of exogenous DNA damage treatment. The “n” numbers of fibers analyzed (pooled from three independent experiments), as well as the mean ± SEM, are indicated on the graphs. P‐values are 9.8 × 10−22 (G) and 1.0 × 10−10 (H).
