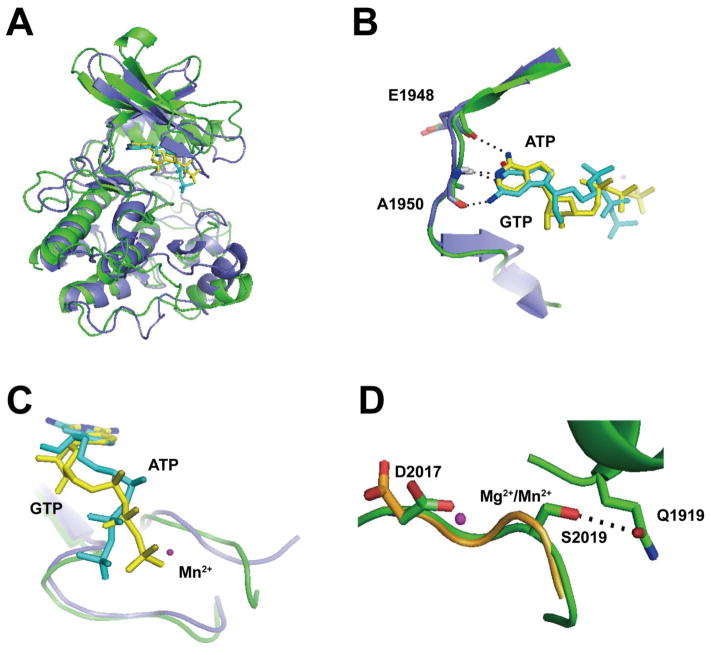
Figure 4. In silico prediction of a novel GTP-bound LRRK2 isoform.
A) Hypothetical model of ATP (yellow) and GTP (cyan)-bound LRRK2 kinase domain based on human MLK3 crystal structure (PDBID: 3DTC, slate) overlaid with the LRRK2 kinase domain constructed from a model based on amoeba ROCO4 crystal structure (PDBID, 4F0F, green). LRRK2 overlaid on either the MLK3 or ROCO4 nucleotide pockets overall resembles each other with an overall root-mean-square deviation of 1.5 Å, and B) in this hypothetical model, the amino group on C6 of ATP could form a hydrogen bond with the main chain oxygen on E1948 (hLRRK2 sequence). The N1 atom could form a hydrogen bond with the main chain amide group of A1950. The N1 atom of GTP could form a hydrogen bond with the main chain amide group on A1950. The amino group on C2 of GTP might form a hydrogen bond with the main chain oxygen of A1950. C) Overlay of bound ATP (yellow) and GTP (cyan) showed the triphosphate tail of GTP must shift away from the bound Mn2+. D) The overlay model of WT (orange) and G2019S (green) LRRK2 kinase domain bound with Mg2+ or Mn2+ showed that in WT LRRK2, D2017 is directed away from the Mn2+. However, in G2019S LRRK2 the Ser2019 residue might form a hydrogen bond with Q1919, necessarily dragging D2017 closer to the Mn2+ atom. Important residues are shown as sticks. Metals are shown as spheres. Atoms involved in key interactions are color coded (blue: nitrogen, red: oxygen). Black dotted lines denote the atoms involved in potential hydrogen bonding.