Abstract
Functional magnetic resonance imaging (fMRI) provides a non-invasive way to investigate brain activity. Recently, convergent evidence shows that the correlations of spontaneous fluctuations between two distinct brain regions dynamically change even in resting state, due to the condition-dependent nature of brain activity. Thus, quantifying the patterns of functional connectivity (FC) in a short time period and changes of FC over time can potentially provide valuable insight into both individual-based diagnosis and group comparison. In light of this, we propose a novel computational method to robustly estimate both static and dynamic spatial-temporal connectivity patterns from the observed noisy signals of individual subject. We achieve this goal in two folds: (1) Construct static functional connectivity across brain regions. Due to low signal-to-noise ratio induced by possible non-neural noise, the estimated FC strength is very sensitive and it is hard to define a good threshold to distinguish between real and spurious connections. To alleviate this issue, we propose to optimize FC which is in consensus with not only the low level region-to-region signal correlations but also the similarity of high level principal connection patterns learned from the estimated link-to-link connections. Since brain network is intrinsically sparse, we also encourage sparsity during FC optimization. (2) Characterize dynamic functional connectivity along time. It is hard to synchronize the estimated dynamic FC patterns and the real cognitive state changes, even using learning-based methods. To address these limitations, we further extend above FC optimization method into the spatial-temporal domain by arranging the FC estimations along a set of overlapped sliding windows into a tensor structure as the window slides. Then we employ low rank constraint in the temporal domain assuming there are likely a small number of discrete states that the brain transverses during a short period of time. We applied the learned spatial-temporal patterns from fMRI images to identify autism subjects. Promising classification results have been achieved, suggesting high discrimination power and great potentials in computer assisted diagnosis.
1 Introduction
In general, resting-state functional connectivity is a set of pair-wise connectivity measurements, each of which describes the strength of co-activity between two regions in human brain. In many group comparison studies, FC obtained from resting-state fMRI shows observable abnormal patterns in patient cohort to understand different disease mechanisms. In clinical practice, FC is regarded as an important biomarker for disease diagnosis and monitoring in various clinical applications such as Alzheimer’s disease [1] and Autism [2].
In current functional brain network studies, Pearson’s correlation on BOLD (Blood Oxygen Level Dependent) signals is widely used to measure the strength of FC between two brain regions [2, 3]. It is worth noting that such correlation based connectivity measure is exclusively calculated based on the observed BOLD signals and fixed for the subsequent data analysis. However, the BOLD signal usually has very poor signal-to-noise ratio and is mixed with substantial non-neural noise and artefacts. Therefore, it is hard for current state-of-the-art methods to determine a good threshold of FC measure which can effectively distinguish real and spurious connections.
For simplicity, many FC characterization methods assume that connectivity patterns in the brain do not change over the course of a resting-state fMRI scan. There is a growing consensus in the neuroimaging field, however, that the spontaneous fluctuations and correlations of signals between two distinct brain regions change with correspondence to cognitive states, even in a task-free environment [4]. Thus, dynamic FC patterns have been investigated recently by mainly using sliding window technique [4, 11–13]. However, it is very difficult to synchronize the estimated dynamic patterns with the real fluctuations of cognitive state, even using advanced machine learning techniques such as clustering [5] and hidden Markov model [6]. For example, both methods have to determine the number of states (clusters) which might work well on the training data but have the potential issue of generality to the unseen testing subjects.
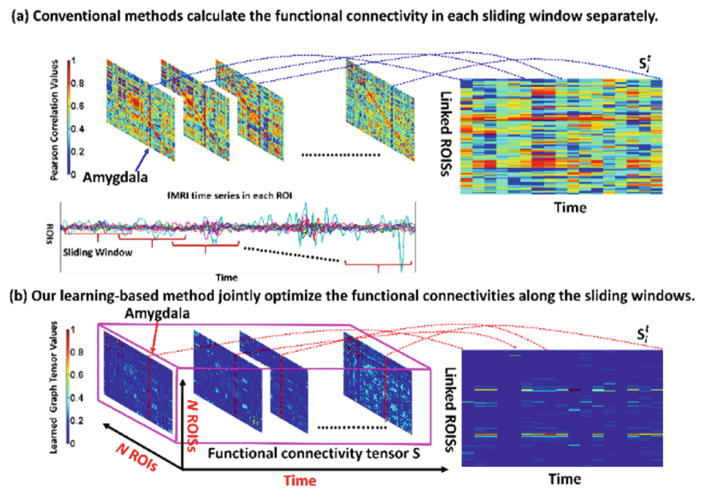
To address above issues, we propose a novel data-driven solution to reveal the consistent spatial-temporal FC patterns from resting-state fMRI image. Our work has two folds. First, we present a robust learning-based method to optimize FC from the BOLD signals in a fixed sliding window. In order to avoid the unreliable calculation of FC based on signal correlations, high level feature representation is of necessity to guide the optimization of FC. Specifically, we apply singular value decomposition (SVD) to the tentatively estimated FC matrix and regard the top ranked eigenvectors are as the high level network features which characterize the principal connection patterns across all brain regions. Thus, we can optimize functional connections for each brain region based on not only the observed region-to-region signal correlations but also the similarity between high level principal connection patterns. In turn, the refined FC can lead to more reasonable estimation of principal connection patterns. Since brain network is intrinsically economic and sparse, sparsity constraint is used to control the number of connections during the joint estimation of principal connection patterns and the optimization of FC. Second, we further extend the above FC optimization framework from one sliding window (capturing the static FC patterns) to a set of overlapped sliding windows (capturing the dynamic FC patterns), as shown in the middle of Fig. 1. The leverage is that we arrange the FCs along time into a tensor structure (pink cubic in Fig. 1) and we employ additional low rank constraint to penalize the oscillatory changes of FC in the temporal domain.
Fig. 1.
The advantage of our learning-based spatial-temporal FC optimization method (bottom) over the conventional method (top) which calculate the FC based on signal correlations. As the trajectory of FC at Amygdala shown in the right, the dynamic FC optimized by our learning-based method is more reasonable than the conventional correlation-based method.
In this paper, we apply our learning-based method to find the spatial-temporal functional connectivity patterns for identifying childhood autism spectrum disorders (ASD). Compared with conventional approaches which simply calculate FC based on signal correlations, more accurate classification results have been achieved in classifying normal control (NC) and ASD subjects by using our learned spatial-temporal FC patterns.
2 Method
2.1 Construct Robust Functional Connectivity
Let xi ∈ ℜW×1 denote the mean BOLD signal calculated in brain region Oi (i = 1, …, N), where W is the length of time course within the sliding window. Conventionally, a N × N connectivity matrix S is used to measure the FCs in the whole brain, where each element sij quantitatively measure the strength of FC between region Oi and Oi (i ≠ j). For convenience, we use si ∈ ℜN×1 denote i-th column in connectivity matrix S, which characters the connections w.r.t. other brain regions. Since the signal-to-noise ratio of observed xi is low, high level feature is of necessity to guide the estimation of connectivity matrix S. To achieve it, we apply singular value decomposition to S and regard the top ranked eigenvectors matrix FK×N =[fi]i=1,…,N as the high level network features, where each fi ∈ ℜK×1 denotes the principal connection pattern on region Oi. Thus, instead of calculating the connectivity sij based on correlation c(xi, xj) between observed BOLD signals xi and xj, we require the optimal connectivity sij should (1) be in consensus with the correlation of low level signals between xi and xj; and (2) be in line with similarity of high level principal connection patterns between fi and fj. To that end, the objective function is defined as:
| (1) |
where r1 is the scalar controlling the strength of connection sparsity for each connection pattern si. In order for robustness, L2 norm is applied to si. Since the estimation of sij and fi are coupled, we propose the following solution to alternative solve sij and fi:
Initialize connectivity matrix by letting sij = c(xi, xj);
Given S, obtain the principal connection pattern fi for each region Oi by applying eigenvalue decomposition to S since S is symmetric. After that, we select the top K eigenevectors.
Fixing fi, we divide the estimation of sij in Eq. (1) into two sub-tasks: (a) Estimate sij without the sparsity constraint. Since the objective function without the L1 norm can be reformulated into a quadratic form, we can use Karush Kuhn Tucker (KKT) [7] algorithm to optimize sij. (b) Make the connection pattern si sparse. The objective function requires the optimized connection pattern si not only sparse but also close to the solution in step 3(a) Standard Alternating Direction Method of Multipliers (ADMM) [7, 8, 14] can be used to solve this sub-task.
Go to step 2 until converge.
Typical optimized connectivity matrix Ŝ is shown in the pink cubic in Fig. 1. Compared to the connectivity matrix by conventional method based on the signal correlation, our learned connectivity matrix is much sparser and it becomes much easier to construct the brain network since a lot of spurious connections have been removed by using the sparsity constraint during optimization.
2.2 Characterize Dynamic Functional Connectivity
Next, we extend our learning based FC estimation method to address the problem of dynamic connectivity in fMRI data. Here, we follow the sliding window technique to obtain T overlapped sliding windows which cover the whole time course. Since we can optimize the connectivity matrix St for each sliding window, we employ a tensor 𝕊 = {St|t=1, …, T} ∈ ℜN×N×T to describe the dynamic connectivity. Similarly, we construct tensor ℂ = {Ct|t = 1, …, T} ∈ ℜN×N×T regarding the dynamic correlation of BOLD signals and tensor 𝔽 = {Ft|t = 1, …, T} ∈ ℜN×N×T regarding the dynamic similarity of principal connection patterns, where and are the N × N matrices in t-th sliding window. Then, we extend the objective function in Eq. (1) to the spatial-temporal domain using tensor analysis:
| (2) |
where and . We use 𝕊(k) denote the unfolding operation to a general tensor 𝕊 along the k-th mode. In our method, we have 𝕊(1) ∈ ℜN2×T and 𝕊(2) ∈ ℜNT×N. Since brain in resting state generally transverses a small number of discrete stages during a short period of time [4], we require the change of connectivity matrix St to be smooth along time. Thus, it is reasonable to apply low rank constraint on 𝕊(1) such that the minimization of ||𝕊(1)||* (nuclear norm of 𝕊(1)) can suppress too rapid FC change in the temporal domain. L1-norm is applied to 𝕊(2) since the brain network within each sliding window is sparse.
Optimization
In order to make the optimization of Eq. (2) tractable, we introduce two dummy variables Z1 and Z2 so that we can solve this problem using ADMM [7, 8]:
| (3) |
Using Lagrangian multipliers, we can remove the equality constraints in Eq. (3) and reformulate Eq. (3) into:
| (4) |
where Λ1 and Λ2 are the N2 × T Largrangian multiplier matrix, and μ1 and μ2 are the penalty parameters. Furthermore, we solve Eq. (4) by alternatively optimize 𝕊, Z1 and Z2 until Eq. (4) converges. The dynamic connectivity matrices St can be optimized by following the Karush Kuhn Tunker (KKT) method in [9]. Standard soft threshold shrinkage method [7] can be used to solve Z1 and Z2.
2.3 Identifying ASD Subject with the Learned Static/Dynamic FC Patterns
Conventional method first calculate the connectivity matrix S within certain window and then extract N dimension node-wise features which describe the connectivity efficiency at each ROI (Region of Interest) [10]. After that, classic SVM (linear kernel and L2 penalty, https://www.csie.ntu.edu.tw/~cjlin/libsvm/) is trained to identify individual ASD subjects. We follow the same approach except we extract the node-wise features from our learned static/dynamic connectivity matrices.
3 Experiment
Image Preprocessing
We conducted various experiments on resting-state fMRI images from both NYU and UM sites in Autism Brain Imaging Data Exchange (ABIDE) database, in order to demonstrate the generality of our method. Specifically, 45 NC and 45 ASD subjects are selected from the NYU site. 74 NC and 57 ASD subjects are selected from UM site. The subjects from NYU site and UM site were scanned for six and ten minutes during resting state, respectively, producing 180 time points and 300 time points at a repetition time (TR) of 2 s. We processed all these data using Data Processing Assistant for Resting-State fMRI (DPARSF) software. Specifically, we remove the first 20 time points and last 20 time points for robustness. After that, we corrected the fMRI images by slice timing and motion correction. Then, we register individual subjects to the standard space, apply the AAL template with 116 ROIs to the subject image domain and compute the mean BOLD signal in each ROI, where conventional method calculate the 116 × 116 connectivity matrix S based on the correlation of mean BOLD signals between any pair of two distinct brain regions.
Experiment Setup
Ten-fold cross validation strategy is used in our experiments. We randomly partition the subjects in the NC and ASD groups into 10 non-overlapping approximately equal size sets. At each subject, we apply our learning-based method to optimize the static/dynamic functional connectivity matrices and extract the node-wise network features. Then, we use one fold for testing and the remaining folds are used for training the SVM. The training subjects are further divided into 5 subsets for another 5-fold inner cross validation to learn the optimal parameters. The optimal principle component range is [6, 8] and it take average 10 s to run one subject.
Evaluation of Learned Static/Dynamic FC Patterns in NC/ASD Classification
We first manually set up the sliding window size which ranges from 20 % to 100 % of the entire time course. In optimizing the dynamic FC pattern, we set the shift of sliding window to 1 TR, in order to fully capture the dynamics of FC. The NC/ASD classification results on UM and NYU dataset are shown in Tables 1 and 2, respectively. In these two tables, ACC represents accuracy and AUC represent the area under an ROC curve. It is clear that SVM using the learned dynamic FC patterns outperforms the conventional correlation based FC patterns and the learned static FC patterns, where using the learned dynamic FC patterns can achieve almost 8 % increase over the correlation-based FC patterns and almost 3 % increase over using the learned static FC patterns.
Table 1.
Accuracy of identifying ASD subjects on UM dataset w.r.t. sliding window size.
| Window size | Perarson correlation | Learned static FC | Learned dynamic FC | |||
|---|---|---|---|---|---|---|
| ACC | AUC | ACC | AUC | ACC | AUC | |
| 10 % | 87.37 ± 6.13 | 93.22 ± 7.53 | 89.23 ± 5.27 | 94.74 ± 6.83 | 92.25 ± 7.21 | 97.31 ± 8.63 |
| 25 % | 84.50 ± 6.51 | 89.71 ± 7.45 | 87.83 ± 4.57 | 92.03 ± 5.81 | 90.35 ± 6.72 | 95.46 ± 7.45 |
| 45 % | 80.81 ± 5.55 | 85.02 ± 6.81 | 84.89 ± 3.78 | 88.91 ± 4.57 | 86.45 ± 5.34 | 91.33 ± 5.81 |
| 60 % | 75.71 ± 8.45 | 78.83 ± 9.51 | 81.71 ± 7.17 | 87.12 ± 8.64 | 83.76 ± 6.52 | 88.42 ± 7.46 |
| 100 % | 68.13 ± 12.41 | 74.32 ± 14.36 | 70.52 ± 11.35 | 75.17 ± 13.11 | 77.85 ± 8.66 | 81.31 ± 9.52 |
Table 2.
Accuracy of identifying ASD subjects on NYU dataset w.r.t. sliding window size.
| Window size | Perarson correlation | Learned static FC | Learned dynamic FC | |||
|---|---|---|---|---|---|---|
| ACC | AUC | ACC | AUC | ACC | AUC | |
| 10 % | 86.59 ± 5.01 | 91.07 ± 6.92 | 88.37 ± 6.07 | 92.36 ± 7.31 | 91.85 ± 5.11 | 96.23 ± 7.24 |
| 25 % | 83.83 ± 5.56 | 88.61 ± 6.12 | 86.89 ± 3.76 | 91.76 ± 4.57 | 89.07 ± 4.71 | 94.64 ± 5.67 |
| 45 % | 77.72 ± 7.45 | 81.57 ± 9.24 | 84.71 ± 6.18 | 89.43 ± 7.17 | 87.25 ± 5.46 | 91.37 ± 6.16 |
| 60 % | 72.96 ± 12.21 | 77.56 ± 13.73 | 78.22 ± 9.14 | 84.56 ± 10.29 | 82.52 ± 7.82 | 86.71 ± 8.62 |
| 100 % | 65.35 ± 12.13 | 70.28 ± 14.52 | 69.33 ± 10.41 | 74.21 ± 11.35 | 75.73 ± 8.27 | 80.16 ± 9.13 |
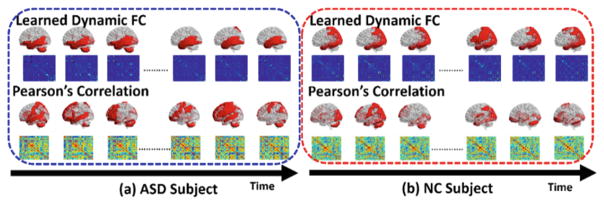
Visualization of Dynamic Function Connectivity Patterns
The learned dynamic functional connectivity matrices {St} for one ASD subject (in blue box) and one NC subject (in red box) are displayed in the top of Fig. 2. For comparison, we also show the corresponding connectivity matrices independently calculated based on the correlation of BOLD signals in the bottom of Fig. 2. It is apparent that our learned dynamic connectivity matrices are much sparser than the counterpart matrices using conventional method. Thus, it becomes much easier to construct functional brain network using threshold based approach. In order to evaluate the dynamic functional transit, we visualize the ROIs connected to Amygdala along time by examining the estimated connectivity matrices since Amygdala is a critical sub-cortical structure related to Autism. As the red masks shown in Fig. 2, the transitions of ROIs connected to Amygdala revealed by our learned dynamic FC patterns are much more consistent than those by conventional correlation based FC patterns, where the transit of connected ROIs is unrealistically fast and random. Specifically, we transpose the connection vector si (i denotes the index for Amygdala here) into column vector and sequentially arrange them into a matrix and visualize the transition of si along time in the right of Fig. 1. From the trajectory of each element in si, it is clear that the dynamic FC optimized by our leaning-based method is more reasonable than the conventional method.
Fig. 2.
Visualization of dynamic functional connectivity matrices by our learning-based method (top) and conventional correlation based method for an ASD subject (a) and a NC subject (b).
Validation of FC Pattern
We also checked the learned fc pattern and found that the connection pattern for vision function regions, such as Lingual gyrus, Cuneus, Parahippocampalgyrus, and for motion function such as Putamen, Globus Pallidus are consistently stable for all subjects. We also found that the learned fc pattern is similar as correlation fc pattern, however, the change is much smoother along time which is consistent with current neuroscience findings.
4 Conclusion
In this work, we propose a novel learning-based method to discover both static and dynamic connectivity patterns from resting-state fMRI data. For static FC estimation, our method optimizes the functional connectivity based on not only the correlation of low level BOLD signals but also the similarity of high level principal components from the link-to-link connectivity patterns. To address the problem of dynamic functional connectivity, we arrange connectivity matrices along time into a tensor structure and apply sparsity to suppress spurious functional connectivities and low rank to avoid unrealistic fast state transition along time. We use our method to obtain dynamic connectivity patterns and apply them to identify ASD subject at individual level, where classification method using our learned dynamic connectivity patterns can improve the ASD identification accuracy with almost 8 % increase conventional correlation-based framework.
References
- 1.Greicius M, Srivastava G, Reiss A, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. PNAS. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 3.van den Heuvel MP, Pol HEH. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20:519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Penna SD, Duyn JH, Glover GH, Gonzalez-Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, de Pasquale F, Sporns O, Walter M, Chang C. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wee CY, Yap PT, Shen D. Diagnosis of autism spectrum disorders using temporally distinct resting-state functional connectivity networks. CNS Neurosci Ther. 2016;22:212–219. doi: 10.1111/cns.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eavani H, Satterthwaite TD, Gur RE, Gur RC, Davatzikos C. Unsupervised learning of functional network dynamics in resting state fMRI. In: Gee JC, Joshi S, Pohl KM, Wells WM, Zöllei L, editors. IPMI 2013. LNCS. Vol. 7917. Springer; Heidelberg: 2013. pp. 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd S, Vandenberghe L. Convex Optimization. Cambridge University Press; Cambridge: 2004. [Google Scholar]
- 8.Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Found Trends Mach Learn. 2011;3:1–122. [Google Scholar]
- 9.Nie F, Wang X, Huang H. Clustering and projected clustering with adaptive neighbors. The 20th International Conference on Knowledge Discovery and Data Mining; 2014. [Google Scholar]
- 10.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Urs B, et al. Dynamic reconfiguration of frontal brain networks during executive cognition in humans. PNAS. 2015;112:11678–11683. doi: 10.1073/pnas.1422487112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heung-Il S, Lee SW, Shen D. A hybrid of deep network and hidden Markov model for MCI identification with resting-state fMRI. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. MICCAI 2015. LNCS. Vol. 9349. Springer; Heidelberg: 2015. pp. 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonardi N, et al. Principal components of functional connectivity: a new approach to study dynamic brain connectivity during rest. Neuroimage. 2013;83:937–950. doi: 10.1016/j.neuroimage.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Lucey S. Convolutional sparse coding for trajectory reconstruction. IEEE Trans Pattern Anal Mach Intell. 2015;37(3):529–540. doi: 10.1109/TPAMI.2013.2295311. [DOI] [PubMed] [Google Scholar]