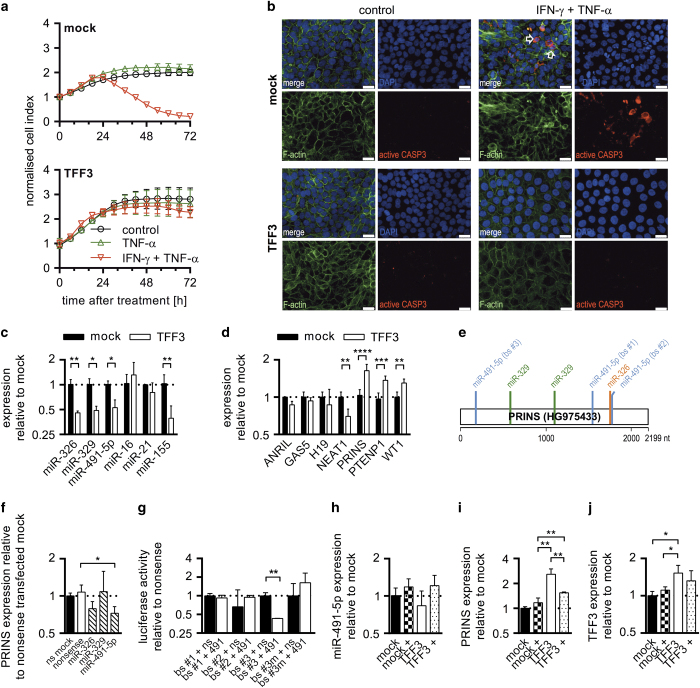
Figure 1.
TFF3 overexpression in HT-29/B6 confers resistance to TNF-α/IFN-γ-induced apoptosis concomitant with dysregulation of apoptosis-related ncRNA. (a) Cell indices of HT-29/B6/TFF3 and mock-transfected colorectal adenocarcinoma cells were determined using xCELLigence analyses over 72 h. Bars show S.D. of at least four replicates. TFF3-overexpressing cells were protected from TNF-α/IFN-γ-induced apoptosis. (b) Apoptosis was evaluated by IF detection of active caspase-3. White arrows indicate apoptotic cells that are positive for active caspase-3 and have fragmented nuclei. Scale bars indicate 25 nm. (c) Expression of miRNAs was determined at day 7 after seeding. White columns show mock controls and black TFF3-overexpressing HT-29/B6 cells. Error bars indicate the S.D. Values were log2 transformed. (d) Expression of lncRNAs was determined at day 7 after seeding. White columns show mock controls and black TFF3-overexpressing HT-29/B6. Error bars indicate the S.D. Values were log2 transformed. (e) Multiple binding sites for miR-326, miR-329 and miR-491-5p were identified in the PRINS sequence (HG975433). (f) HT-29/B6/TFF3 were transfected with miR-326, miR-329, miR-491-5p (hatched columns) or nonsense (ns) controls (white column) and expression of PRINS was compared with ns-transfected mock (black column) 24 h post treatment. Error bars indicate the S.D. Values were log2 transformed. (g) Luciferase reporter gene assays identified miR-491-5p to target binding site (bs) #3. The mutated site (bs #3 m) served as a control. Samples were taken at 24 h after transfection. Black columns show ns-transfected controls. Error bars indicate the S.D. Values were log2 transformed. (h–j) Bar diagrams show the expression of miR-491-5p, PRINS and TFF3 in TNF-α/IFN-γ-treated (+) HT-29/B6/TFF3 (dotted) and mock cells (squares) compared with untreated mock controls (black) and HT-29/B6/TFF3 (white). Error bars indicate the S.D. Values were log2 transformed. Columns show means of at least three biological replicates and bars indicate S.D. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001, unpaired t-test.