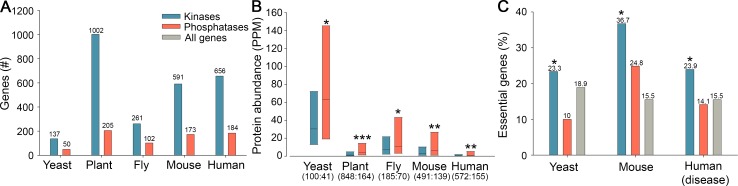
Fig 1. Differences in gene numbers, protein abundance and essentiality between kinases and phosphatases are conserved across eukaryotic lineages.
A. Kinase-coding genes are more abundant than phosphatase-coding genes in five eukaryotic genomes. B. Phosphatase proteins are significantly more abundant than kinase proteins in five eukaryotic proteomes. Box plots show the values at the first, second and third quartiles. In parenthesis per organism are the numbers of kinases and phosphatases for which data were available. Median values for kinases and phosphatases, and the respective Mann-Whitney p-values, were as follows: Yeast 30.4, 63, p = 0.008; plant 1.3, 4, p<10−10, fly 7.5, 10.6, p = 0.03; mouse 2.8, 6.3, p = 9*10−4; human 0.33, 0.65, p = 5*10−4. C. The fraction of phosphatases that are essential for survival in yeast and mouse, or are associated with genetic disease in human, is significantly smaller than the fraction of kinases (yeast p = 0.03, mouse p = 0.0022, human p = 0.0023; Fisher exact test). Yeast = Saccharomyces cerevisiae; Plant = Arabidopsis thaliana; Fly = Drosophila melanogaster; Mouse = Mus musculus; Human = Homo sapiens. *** indicates p<10−6; ** indicates p<10−3; * indicates p<0.05. Numbers above bars indicate Y-axis values.