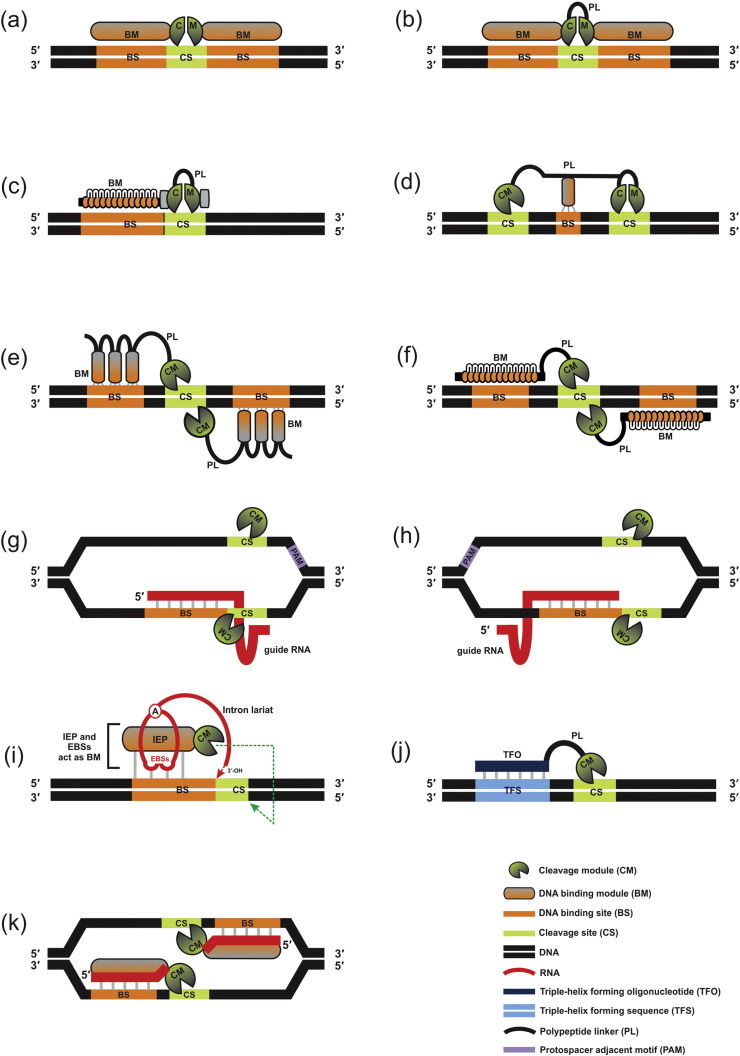
Fig. 1.
Examples of programmable genome editing tools. (a) Single-motif LAGLIDADG homing endonucleases, (b) double-motif LAGLIDADG homing endonucleases, (c) megaTAL, (d) MegaTev, (e) zinc-finger nucleases (ZFN), (f) transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated proteins (Cas) systems using (g) Cas9 or (h) Cpf1, (i) targetrons, (j) triplex-forming oligonucleotide (TFO) nucleases, and (k) structure-guided nucleases (SGNs). EBS = exon-binding site; IEP = intron-encoded protein. The nuclease domain of FokI is used to engineer ZNFs, TALENs, and SGNs. Elements of this figure have been adapted from Hafez et al. [44] NRC Research Press License number: 3981970186164.