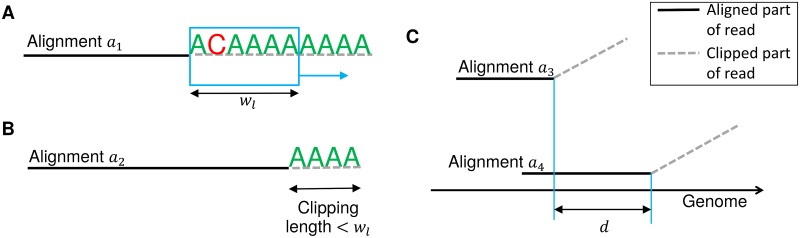
Fig 2. Identification of candidate poly(A) sites.
(A) For each alignment, a sliding window of length wl is shifted along the clipped part of the read sequence and the fraction of A’s (or T’s depending on strandedness of sequencing) is calculated within each window. In this example, the fraction is 5/6 = 0.83 for the first two windows and 6/6 = 1 for all subsequent windows. Thus, at least one window contains ≥ c1 = 1 A’s and none has < c2 = 0.7 A’s and this is used as a candidate poly(A) site. (B) In this example, the clipping length is shorter than wl. Accordingly, the window approach cannot be used and all clipped nucleotides are required to be A’s or T’s to predict a candidate poly(A) site, which is the case here. (C) Alignments a3 and a4 are considered pairwise overlapping as they are clipped at the same end (dashed lines) and the distance d between the start of clipping is smaller than the read length.