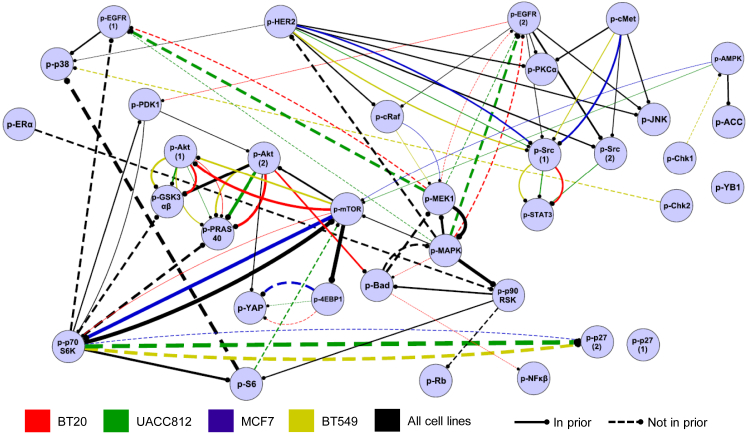
Figure 5.
Context-Specific Signaling Networks Reconstructed Using a Machine Learning Approach
Data for 35 phosphoproteins were analyzed using a machine learning approach based on interventional dynamic Bayesian networks, integrating also known biology in the form of a prior network (Figure S4). This gave a set of scores (edge probabilities) for each possible edge in each (cell line, stimulus) context (see STAR Methods). For each cell line, a summary network was obtained by averaging edge probability scores across the eight stimulus-specific networks for that cell line. Edge color denotes cell line. Only edges with average probabilities greater than 0.2 are shown. A black edge indicates an edge that appears (i.e., is above the 0.2 threshold) in all four cell lines. Edge thickness is proportional to the average edge probability (average taken across all 32 contexts for black edges). Solid or dashed edges were present or not present in the prior network, respectively. Edges are directed with the child node indicated by a circle. Edge signs are not reported; the modeling approach does not distinguish between excitatory and inhibitory causal effects. Full node names, including phosphorylation sites, are provided in Table S4. Network visualized using Cytoscape (Shannon et al., 2003). See also Table S3.