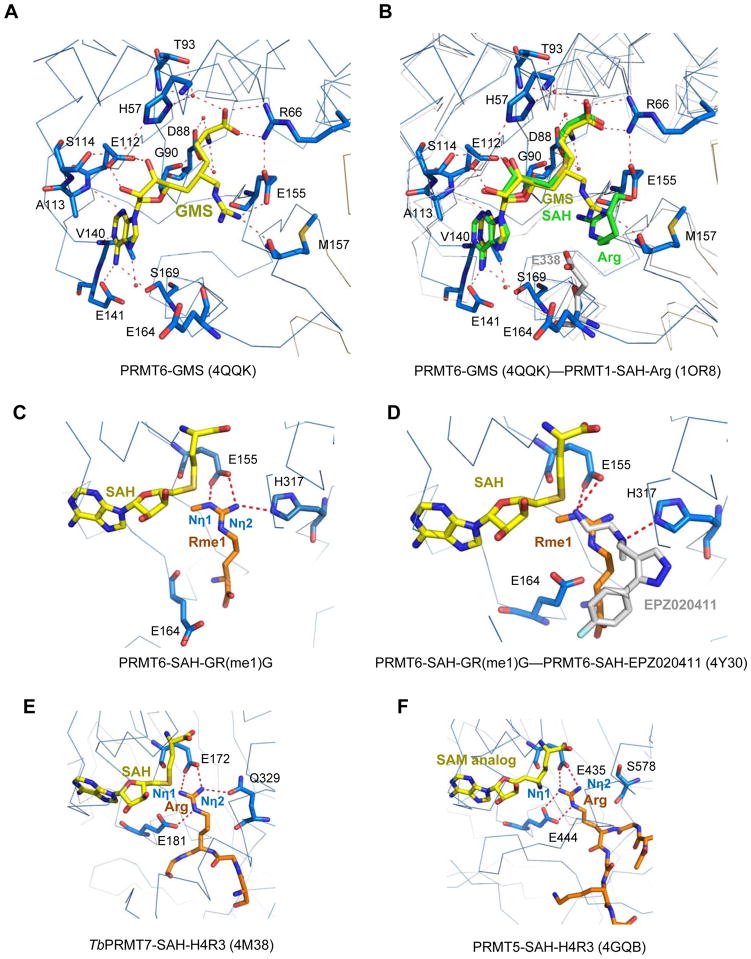
Figure 5.
Structural comparison of arginine methyltransferases in complex with different ligands. (A) Detailed interactions between PRMT6 and the bi-substrate inhibitor GMS (PDB: 4QQK, this work). GMS is shown in a stick model and colored in yellow. The GMS interaction residues in PRMT6 are shown in stick models. (B) Superposition of the PRMT6-GMS (PDB: 4QQK, this work), PRMT1-SAH-arginine (PDB: 1OR8) and PRMT3-SAH (PDB: 2FYT). PRMT6, PRMT1 and PRMT3 have very conserved structures. For clarity, only PRMT6 (blue) and PRMT1 (grey) are shown in ribbons. GMS from the PRMT6 structure is shown in a yellow stick model. SAH and the substrate arginine residue from the PRMT1-SAH-arginine structure are shown in green stick models. For PRMT3 structure, only the second glutamic acid residue E338 from the double-E loop of PRMT3 is shown in a grey stick model. (C) Detailed interactions between PRMT6 and the mono-methyl arginine of the GR(me1)G peptide. E164 is superimposed from the PRMT6-SAH-EPZ020411 structure. (D) Superposition of the PRMT6-SAH-GR(me1)G and PRMT6-SAH-EPZ020411 (PDB: 4Y30). SAH, mono-methyl arginine, EPZ020411 and the interacting residues in PRMT6 are shown in stick models and colored in yellow, orange, grey, and blue, respectively. (E) Detailed interactions between TbPRMT7 and the substrate arginine residue (PDB: 4M38). (F) Detailed interactions between PRMT5 and the substrate arginine residue (PDB: 4GQB).