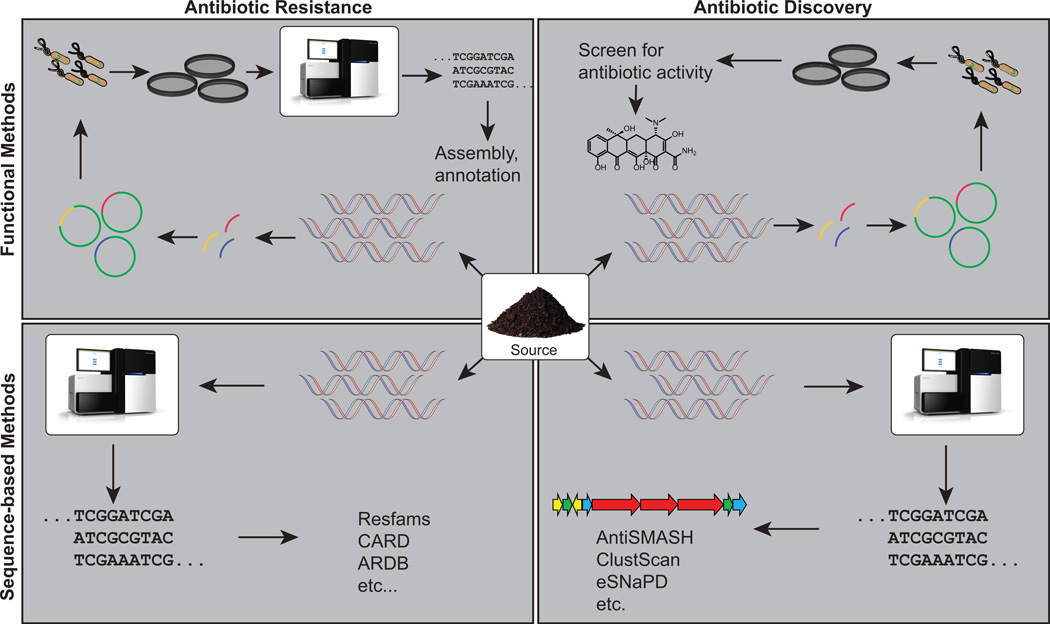
Figure 2.
An overview of function- and sequence-based methods for the discovery of antimicrobials and antimicrobial resistance from microbiomes. DNA is isolated from source material (center), which can then be mined for antimicrobial resistance genes (left) or antimicrobial biosynthetic pathways (right). Functional metagenomic methods (top) typically entail shotgun cloning metagenomic DNA into an expression vector and selecting for a desired phenotype (e.g., antimicrobial resistance or antimicrobial activity). Sequence-based methods (bottom) customarily involve sequencing metagenomic DNA and annotating sequences using general or function-specific databases.